Muscling RNA polymerase off the DNA
Researchers elucidate a unique molecular mechanism for efficient gene expression in pathogenic bacteria
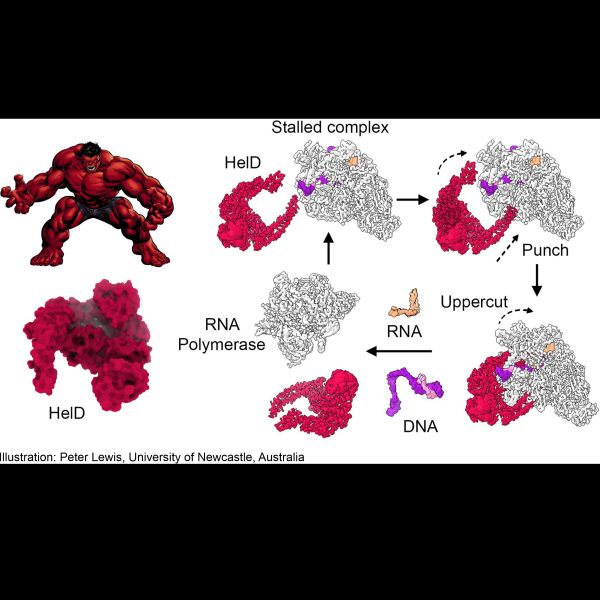
Three international research teams, including a group from the University of Newcastle, find that a motor protein, called HelD, acts like a “molecular bully” to pry the central enzyme of transcription, RNA polymerase, away from the DNA template, setting it free for the continued production of genetic messages. The findings uncover molecular mechanisms important for the efficient growth of pathogenic bacteria, and for their ability to escape the immune system by entering dormant states. The new insights were published in the journal Nature Communications.
Life depends on the flow of the genetic information from DNA to RNA to proteins. In all forms of life, the first step of this gene expression process, transcription of DNA into RNA, is carried out by a complex enzyme, called RNA polymerase.
Being unicellular organisms, bacteria often experience fast-changing environments, for example when they infect their hosts. To survive and reproduce, they have to quickly respond to these environmental changes by adjusting their gene expression programs. Bacteria meet this task, first and foremost, by adjusting their transcription programs. To this end, RNA polymerase has to work optimally, going swiftly through multiple rounds of transcription of the genes on DNA. However, RNA polymerase frequently gets stuck on the DNA, unable to transcribe further or to let go of the template. If left unattended, stalled RNA polymerases compromise continued gene expression and endanger the integrity of the entire genome, because they represent obstacles to the molecular machinery that constantly duplicates the DNA. How RNA polymerases can be efficiently rescued from such trapped states has so far remained enigmatic.
Three research teams around the world have now elucidated how some bacteria solve this fundamental problem - these bugs enlist a molecule called HelD, that has powerful "arms" to reach deep into RNA polymerase. HelD uses these arms to pry the enzyme wide open and to swipe away all bound nucleic acids, reminiscent of a well-muscled comic book hero (see Figure). To elucidate this astounding, "brute force" mechanism of RNA polymerase recovery, all three groups used a technique called single-particle cryogenic electron microscopy, a powerful approach for which the Nobel Prize in Chemistry was awarded in 2017, that can resolve the 3D structures of macromolecular complexes at high resolution.
An Australian team, headed by Peter Lewis (University of Newcastle) and Gökhan Tolun (University of Wollongong), studied the HelD complex of RNA polymerase from an important model bacterium, Bacillus subtilis, which is closely related to dangerous human pathogens, such as Bacillus anthracis (the causative agent of anthrax) and Clostridium difficile (which causes lethal pseudomembranous colitis and diarrhea).
The Australian group also elucidated the first structure of Bacillus subtilis RNA polymerase in the process of transcription. "Together, these structures directly illustrate the dramatic opening of the enzyme caused by HelD", says Prof. Lewis. A Czech consortium led by Libor Krasny and Jan Dohnalek (Czech Academy of Sciences) discovered a variant of the same mechanism in Mycobacteria, which cause tuberculosis and other devastating diseases. "The HelD molecule in Mycobacteria is endowed with an additional spring-loaded arm that can be launched into RNA polymerase to remove the bound nucleic acids. Our structures also show how HelD first grabs RNA polymerase with one arm, then uses the others to force open the enzyme and remove nucleic aicds" notes Dr. Krasny.
Finally, a consortium of researchers from Germany, the USA and Finland, led by Markus Wahl (Freie Universität Berlin), elucidated how a particular building block of RNA polymerase helps HelD in its action in some bacteria. These researchers also provided evidence that under conditions of low energy supply, HelD may promote packaging of RNA polymerase in an inactive form.
"Interestingly, HelD does not invest the universal energy currency of cells, ATP, to force RNA polymerase open; instead, HelD needs ATP-derived energy to let RNA polymerase go again" explains Prof. Wahl.
Related news
- Engineering Students Showcase Innovation at Final Year Project Event
- Former Australian Prime Minister honoured at University of Newcastle graduations
- Advancing Human-Agent Collaboration Through Agentic AI
- Breaking barriers: First doctors graduate from equity pathway
- Translating compassion: a linguist's commitment to social inclusion
The University of Newcastle acknowledges the traditional custodians of the lands within our footprint areas: Awabakal, Darkinjung, Biripai, Worimi, Wonnarua, and Eora Nations. We also pay respect to the wisdom of our Elders past and present.