Professor Matt Dun
Professor
School of Biomedical Sciences and Pharmacy
- Email:matt.dun@newcastle.edu.au
- Phone:(02) 4921 5693
Career Summary
Biography
PROFESSOR MATT DUN - BIOMEDICAL SCIENTIST AND 'DIPG DAD'
Professor Matt Dun is a National Health and Medical Research Council (NHMRC) Emerging Leadership Fellow (2020-2024) and a Defeat DIPG Chadtough New Investigator (2020-2021). Matt is a medical biochemist and Professor of Paediatric Haematology and Oncology Research the University of Newcastle, Australia, and a Deputy Director of the HMRI Precision Medicine Research Program. His position and research has been supported by state and/or national funding bodies continuously since 2012, as well as supplemented by philanthropic and industry contributors throughout.
Decorated by more than 30 national and international awards, Matt achieved his PhD at The University of Newcastle and Hunter Medical Research Institute by publication in 2012. Soon after, he refocused his research from reproductive cell biology and biochemistry, to the fields of medical biochemistry, cancer cell biology and his now specialty - proteomics, proteogenomics, cancer cell biology, brain cancer and leukaemia research.
International post-doctoral training (2013/2014) with field-leading experts in phosphoproteomics (Prof Larsen at SDU) and leukaemia signalling (Prof Cools at KU Leuven), set Matt on a path to establishing his own research laboratory. With his research vision strongly aligned with the HMRI's Precision Medicine Research Program , Matt formed the Cancer Signalling Research Group (CSRG) based at the University of Newcastle (UON) in 2014. This was facilitated (and later enhanced) via his securement of successive Cancer Institute NSW Early Career Fellowships (2014-2016, 2017-2019), NHMRC Investigator Grant (2020-2025), Defeat DIPG Chadtough New Investigator Grant (2020-2021) and successful Cancer Institute NSW and NHMRC Equipment Grants to establish a high-resolution mass spectrometry platform at the UON.
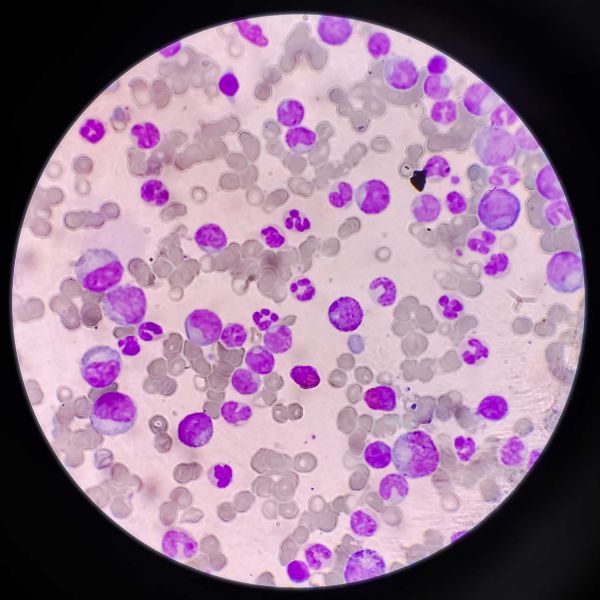
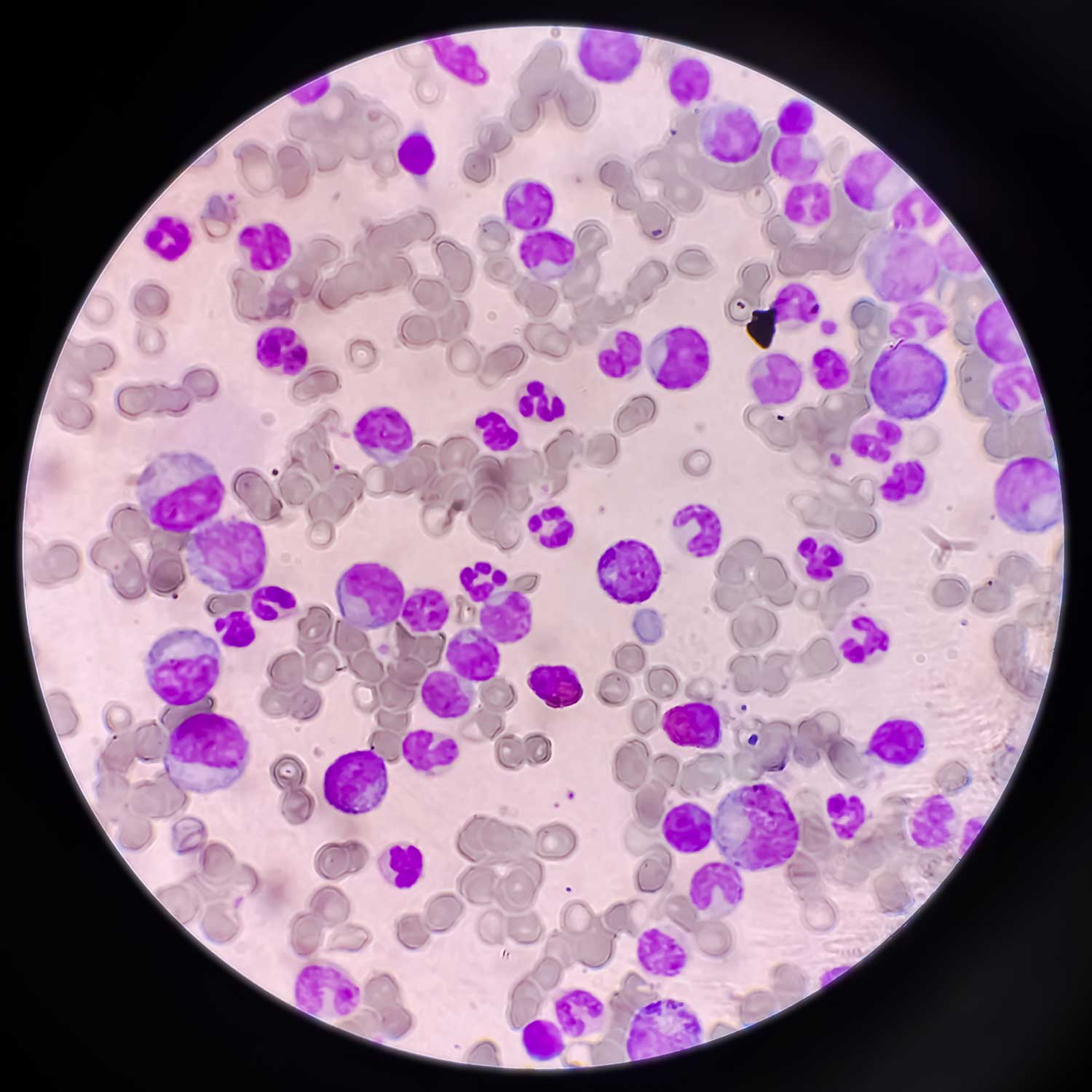
Recently bestowed as the NSW Premier’s Award for Outstanding Cancer Research Fellow of 2019, a Yung Tall Poppy in 2020 and The College of Health, Medicine and Wellbeing Midcareer Researcher of the year 2022, Matt guides the CSRG team of staff and students in fields of paediatric leukaemia, and brain cancer research. The group employ sophisticated phosphoproteomic techniques to characterise the cellular signalling pathways dysregulated by the genetics individualities of a patient’s cancer. This profiling strategy attempts to identify novel treatment targets and drug combinations to improve survival. A member the CNS Tuour Group of the Australian and New Zealand Childhood Haematology and Oncology Group (ANZCHOG) of the Australian & New Zealand Children’s Haematology/Oncology Group (ANZCHOG) and the Australian Leukaemia and Lymphoma Group (ALLG), Matt’s latest collaboration with local investigators (Verrills, Enjeti, Lee) tests a new therapy for acute myeloid leukaemia (AML), and aims to identify the influences of the epigenome of the clonal evolution of AML; both awarded NHMRC Ideas Grant funding from 2020.
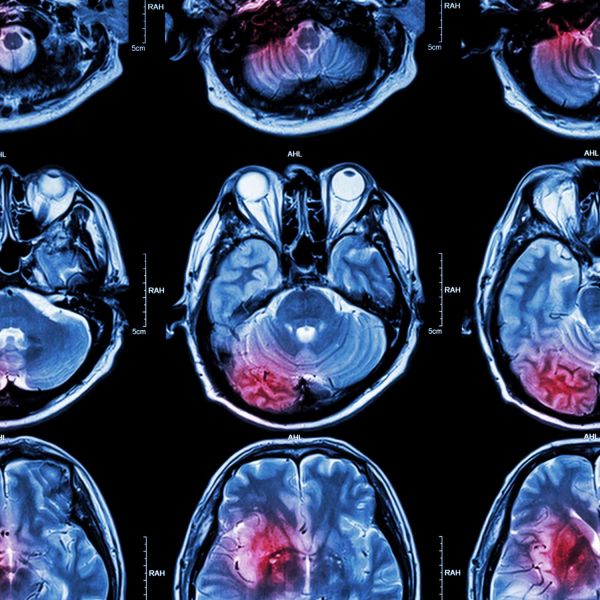
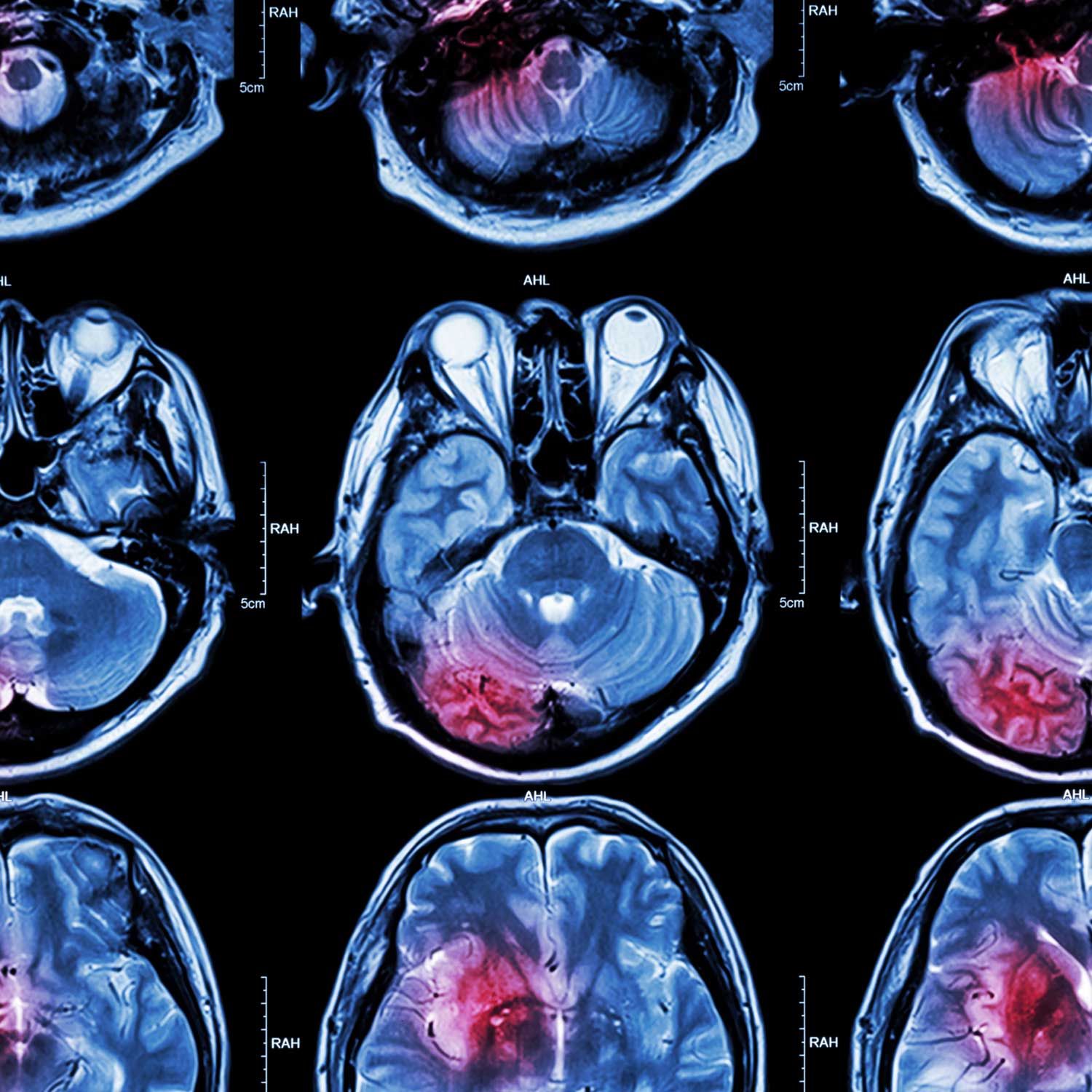
Although initially focused on blood cancers, Matt’s research faced an unexpected shift in focus in 2018, when his then 2-year-old daughter Josephine, was diagnosed with Grade IV diffuse intrinsic pontine glioma (DIPG). Also known as diffuse midline glioma (DMG), Matt was struck by the lack of scientific knowledge regarding the pathophysiology of the disease, and by the stark absence of treatments for DIPG patients - so he set about creating his own program of DIPG research. Guided by those at the forefront of worldwide DIPG research (Monje, Nazarian, Mueller), and self-funded (thanks to the establishment of his own charity ‘RUN DIPG’), Matt’s endeavours have led to the first, high-resolution, quantitative proteomic analysis of the disease.
The esteem of Matt’s DIPG work was highlighted via his invited presentation at the International DIPG Symposium, August 2019 and 2021, and the award of NHMRC Investigator and Defeat DIPG Chadtough New Investigator fellowships. In 2020, Matt has joined (by invitation) the 'Preclinical Working Group' of the Pacific Neuro Oncology Consortium (PNOC) known as 'DMG-ACT'. Basic and discovery scientists worldwide share their latest preclinical DIPG data with clinical collaborators whom lead DIPG/DMG clinical trials - greatly enhancing opportunities for research impact and translation to improved patient outcomes. In 2022, Matt was invited to present the research of his group around the world including an Oral Presentation at the 2022 annual Society for Neuro-Oncology Conference held in Tampa USA.
Dr Dun has 80 refereed journal articles and book chapters, with an accompanying 2500 citations and H-index of 27. In his short career, Matt has been a Chief Investigator on three NHMRC grants including CIA of the Investigator Grant fellowship scheme commencing 2020. Matt’s total research income since PhD award in 2012 is >$13.8 million. Matt is an invited editor to Frontiers of Oncology, Section Editor of Neuro-Oncology and is a Peer Review panelist for the NHMRC Investigator Grants 2019 - .
AWARDS AND MERITS
2022 Mid-Career Researcher of the Year – College of Health, Medicine, and Wellbeing, The University of Newcastle.2022 Directors Mid-Career Researcher of the Year Finalist – Hunter Medical Research Institute.
2022 Big Hero Award – ChadTough Defeat DIPG Foundation, Washington D.C.
2022 Top Ranking Abstract, Plenary Presentation – Society of Neurooncology (SNO) meeting, Tampa Florida.
2020 Young Tall Poppy Science Award – Australian Institute of Policy and Science.
2020 Kellerman Award for Outstanding Contribution to Medical Biochemistry – University of Newcastle.
2020 Achievement in Paediatric Research – Hunter Children’s Research Foundation.
2020-25 Investigator Grant Emerging Leader 1 – National Health and Medical Research Council.
2019 NSW Premier’s Award for Outstanding Research Fellow 2019 – Cancer Institute NSW.
2017 Directors ECR of the Year Finalist – Hunter Medical Research Institute.
2017 Early Career Research Fellowship – Cancer Institute NSW.
2017 HMRI Director’s ECR of the Year Finalist – Hunter Medical Research Institute.
2016 NSW Premier’s Award Cancer Research Fellow of the Year, Finalist – Cancer Institute NSW.
2015 FEBS Journal Best Overall Poster Presentation – Cell Signalling and its Therapeutic Implications.
2015 NSW Premier’s Award Cancer Research Fellow of the Year, Finalist – Cancer Institute NSW.
2014 Early Career Research Fellowship – Cancer Institute NSW.
2013 NSW Office for Health and Medical Research Postdoctoral Award for Excellence in Medical Research – The Australian Society of Medical Research.
2013 The Vice Chancellors Annual Award for Research Higher Degree Excellence – The University of Newcastle.
2013 Translational Cancer Research Fellow – Cancer Institute NSW.
2012 The Deputy Vice Chancellor of Research Award for Research High Degree Excellence – The University of Newcastle.
2012 Best Oral Presentation – Hunter Medical Research Institute Cancer Research Symposium.
2012 The Sydney Catalyst Translational Cancer Research Centre Best Cancer Related Poster – The Garvan Signalling Symposium.
2011 Award for Excellence in Postgraduate Achievement – The University of Newcastle.
2011 2nd Prize for Best Oral Presentation – The Australian and New Zealand Society of Developmental Biology.
2011 Excellence in Research Nomenclature – Society for the Study of Reproduction Meeting, Portland Oregon, USA.
2010 Best Poster Presentation – International Symposium on Spermatology, Okinawa, Japan.
2010 Best Second Year Ph.D. Student Oral Presentation, Research High Degree Conference – The University of Newcastle.
2009 Young Investigator Finalist – The Society of Reproductive Biology.
2009 Best Abstract Presentation, Research High Degree Conference – The University of Newcastle.
2008 Oozoa Award Best Student Oral Presentation – The Society of Reproductive Biology.
PROFESSIONAL MEMBERSHIP
2023- Board - College of Health, Medicine, and Wellbeing, The University of Newcastle
2023- Member - ANZCHOG CNS Tumour Group
2021- Deputy Director - HMRI Precision Medicine Research Program
2020- Member - Society for Neuro-Oncology
2019 - Executive - Preclinical Working Group Pacific Neuro Oncology Consortium DMG-ACT
2019 - Ordinary Member - Australian & New Zealand Children’s Haematology/Oncology Group
2019 - Member - The Brain Foundation
2015 - Member - Australasian Proteomics Society
2013 - Member - The American Association of Cancer Research
2012 - Member - Australian Leukaemia and Lymphoma Group
2012 - Member - The Australian Society for Medical Research
2008-12 Member - The Society for Reproductive Biology
Qualifications
- PhD (Biological Science), University of Newcastle
- Bachelor of Biotechnology, University of Newcastle
- Bachelor of Biotechnology (Honours), University of Newcastle
Keywords
- Anti-Cancer Drug Targeting
- Biochemical Assays
- Biochemistry
- Cancer
- Chemical Proteomics
- Leukaemia
- Medical Biochemistry
- Nucleic Acids
- Phosphoproteomics
- Protein Chemistry
- Proteomics
- acute lymphoblastic leukaemia (ALL)
- acute myeloid leukaemia (AML)
- cell signalling
- diffuse intrinsic pontine glioma (DIPG)
- diffuse midline glioma (DMG)
- mass spectrometry
Fields of Research
| Code | Description | Percentage |
|---|---|---|
| 321106 | Haematological tumours | 30 |
| 321111 | Solid tumours | 30 |
| 320506 | Medical biochemistry - proteins and peptides (incl. medical proteomics) | 40 |
Professional Experience
UON Appointment
| Title | Organisation / Department |
|---|---|
| Associate Professor | University of Newcastle School of Biomedical Sciences and Pharmacy Australia |
Academic appointment
| Dates | Title | Organisation / Department |
|---|---|---|
| 1/12/2013 - | Cancer Instititue NSW Early Career Fellowship | Cancer Instititue NSW School of Biomedical Sciences and Pharmacy, Medical Biochemistry Australia |
| 1/1/2013 - | Conference Chair - The Australian Society for Medical Research, Satellite Scientific Meeting Convenor | The Australian Society for Medical Research |
| 1/10/2011 - 1/12/2013 | HMRI Cancer Research Program Chemical Proteomics Postdoctoral Fellow | University of Newcastle HMRI Cancer Research Program Australia |
Membership
| Dates | Title | Organisation / Department |
|---|---|---|
| 1/1/2013 - | Membership - The Australian Society for Medical Research | The Australian Society for Medical Research |
Publications
For publications that are currently unpublished or in-press, details are shown in italics.
Highlighted Publications
| Year | Citation | Altmetrics | Link | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2021 |
Duchatel RJ, Mannan A, Woldu AS, Hawtrey T, Hindley PA, Douglas AM, et al., 'Preclinical and clinical evaluation of German-sourced ONC201 for the treatment of H3K27M-mutant diffuse intrinsic pontine glioma.', Neuro-oncology advances, 3 vdab169 (2021) [C1]
|
Nova | |||||||||
Chapter (4 outputs)
| Year | Citation | Altmetrics | Link | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2021 |
Nixon B, Cafe SL, Bromfield EG, De Iuliis G, Dun M, 'Capacitation and Acrosome Reaction: Histochemical Techniques to Determine Acrosome Reaction', Manual of Sperm Function Testing in Human Assisted Reproduction, Cambridge University Press, Cambridge 81-92 (2021)
|
||||||||||
| 2019 |
Gould T, Jamaluddin M, Petit J, King SJ, Nixon B, Scott R, et al., 'Finding Needles in Haystacks: The Use of Quantitative Proteomics for the Early Detection of Colorectal Cancer', Advances in the Molecular Understanding of Colorectal Cancer, IntechOpen, Switzerland 1-32 (2019) [B1]
|
Nova | |||||||||
| 2017 |
Nixon B, Dun MD, Aitken RJ, 'Proteomic Analysis of Human Spermatozoa', Immune Fertility: Impact of Immune Reactions on Human Fertility, Springer Nature, Switzerland 3-22 (2017) [B1]
|
Nova | |||||||||
| 2010 |
Dun MD, Mitchell LA, Aitken RJ, Nixon B, 'Sperm-zona pellucida interaction: Molecular mechanisms and the potential for contraceptive intervention', Fertility Control, Springer, Berlin 139-178 (2010) [B1]
|
Nova | |||||||||
| Show 1 more chapter | |||||||||||
Journal article (92 outputs)
| Year | Citation | Altmetrics | Link | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2024 |
Calvert L, Martin JH, Anderson AL, Bernstein IR, Burke ND, De Iuliis GN, et al., 'Assessment of the impact of direct in vitro PFAS treatment on mouse spermatozoa.', Reprod Fertil, 5 (2024) [C1]
|
Nova | |||||||||
| 2024 |
Duchatel RJ, Jackson ER, Parackal SG, Kiltschewskij D, Findlay IJ, Mannan A, et al., 'PI3K/mTOR is a therapeutically targetable genetic dependency in diffuse intrinsic pontine glioma.', J Clin Invest, 134 (2024) [C1]
|
Nova | |||||||||
| 2024 |
Murray HC, Miller K, Dun MD, Verrills NM, 'Pharmaco-phosphoproteomic analysis of cancer-associated KIT mutations D816V and V560G.', Proteomics, e2300309 (2024) [C1]
|
||||||||||
| 2024 |
Skerrett-Byrne DA, Stanger SJ, Trigg NA, Anderson AL, Sipila P, Bernstein IR, et al., 'Phosphoproteomic analysis of the adaption of epididymal epithelial cells to corticosterone challenge', ANDROLOGY, [C1]
|
||||||||||
| 2024 |
de la Nava D, Ausejo-Mauleon I, Laspidea V, Gonzalez-Huarriz M, Lacalle A, Casares N, et al., 'The oncolytic adenovirus Delta-24-RGD in combination with ONC201 induces a potent antitumor response in pediatric high-grade and diffuse midline glioma models.', Neuro Oncol, (2024) [C1]
|
||||||||||
| 2024 |
Koschmann C, Al-Holou WN, Alonso MM, Anastas J, Bandopadhayay P, Barron T, et al., 'A road map for the treatment of pediatric diffuse midline glioma.', Cancer Cell, 42 1-5 (2024) [C1]
|
Nova | |||||||||
| 2023 |
Jackson ER, Persson ML, Fish CJ, Findlay IJ, Mueller S, Nazarian J, et al., 'A review of the anti-tumor potential of current therapeutics targeting the mitochondrial protease ClpP in H3K27-altered, diffuse midline glioma.', Neuro Oncol, (2023) [C1]
|
||||||||||
| 2023 |
Sun CX, Daniel P, Bradshaw G, Shi H, Loi M, Chew N, et al., 'Generation and multi-dimensional profiling of a childhood cancer cell line atlas defines new therapeutic opportunities.', Cancer Cell, 41 660-677.e7 (2023) [C1]
|
Nova | |||||||||
| 2023 |
Thomas BC, Staudt DE, Douglas AM, Monje M, Vitanza NA, Dun MD, 'CAR T cell therapies for diffuse midline glioma.', Trends Cancer, 9 791-804 (2023) [C1]
|
Nova | |||||||||
| 2023 |
Vitanza NA, Wilson AL, Huang W, Seidel K, Brown C, Gustafson JA, et al., 'Intraventricular B7-H3 CAR T Cells for Diffuse Intrinsic Pontine Glioma: Preliminary First-in-Human Bioactivity and Safety.', Cancer Discov, 13 114-131 (2023) [C1]
|
||||||||||
| 2023 |
Jackson ER, Duchatel RJ, Staudt DE, Persson ML, Mannan A, Yadavilli S, et al., 'ONC201 in combination with paxalisib for the treatment of H3K27-altered diffuse midline glioma.', Cancer research, CAN-23-0186 (2023) [C1]
|
Nova | |||||||||
| 2023 |
Messinger D, Harris MK, Cummings JR, Thomas C, Yang T, Sweha SR, et al., 'Therapeutic targeting of prenatal pontine ID1 signaling in diffuse midline glioma', Neuro-oncology, 25 54-67 (2023) [C1] BACKGROUND: Diffuse midline gliomas (DMG) are highly invasive brain tumors with rare survival beyond two years past diagnosis and limited understanding of the mechanism behind tum... [more] BACKGROUND: Diffuse midline gliomas (DMG) are highly invasive brain tumors with rare survival beyond two years past diagnosis and limited understanding of the mechanism behind tumor invasion. Previous reports demonstrate upregulation of the protein ID1 with H3K27M and ACVR1 mutations in DMG, but this has not been confirmed in human tumors or therapeutically targeted. METHODS: Whole exome, RNA, and ChIP-sequencing was performed on the ID1 locus in DMG tissue. Scratch-assay migration and transwell invasion assays of cultured cells were performed following shRNA-mediated ID1-knockdown. In vitro and in vivo genetic and pharmacologic [cannabidiol (CBD)] inhibition of ID1 on DMG tumor growth was assessed. Patient-reported CBD dosing information was collected. RESULTS: Increased ID1 expression in human DMG and in utero electroporation (IUE) murine tumors is associated with H3K27M mutation and brainstem location. ChIP-sequencing indicates ID1 regulatory regions are epigenetically active in human H3K27M-DMG tumors and prenatal pontine cells. Higher ID1-expressing astrocyte-like DMG cells share a transcriptional program with oligo/astrocyte-precursor cells (OAPCs) from the developing human brain and demonstrate upregulation of the migration regulatory protein SPARCL1. Genetic and pharmacologic (CBD) suppression of ID1 decreases tumor cell invasion/migration and tumor growth in H3.3/H3.1K27M PPK-IUE and human DIPGXIIIP* in vivo models of pHGG. The effect of CBD on cell proliferation appears to be non-ID1 mediated. Finally, we collected patient-reported CBD treatment data, finding that a clinical trial to standardize dosing may be beneficial. CONCLUSIONS: H3K27M-mediated re-activation of ID1 in DMG results in a SPARCL1+ migratory transcriptional program that is therapeutically targetable with CBD.
|
Nova | |||||||||
| 2023 |
Germon ZP, Sillar JR, Mannan A, Duchatel RJ, Staudt D, Murray HC, et al., 'Blockade of ROS production inhibits oncogenic signaling in acute myeloid leukemia and amplifies response to precision therapies.', Sci Signal, 16 eabp9586 (2023) [C1]
|
Nova | |||||||||
| 2023 |
Humphries S, Bond DR, Germon ZP, Keely S, Enjeti AK, Dun MD, Lee HJ, 'Crosstalk between DNA methylation and hypoxia in acute myeloid leukaemia', Clinical Epigenetics, 15 (2023) [C1] Background: Acute myeloid leukaemia (AML) is a deadly disease characterised by the uncontrolled proliferation of immature myeloid cells within the bone marrow. Altered regulation ... [more] Background: Acute myeloid leukaemia (AML) is a deadly disease characterised by the uncontrolled proliferation of immature myeloid cells within the bone marrow. Altered regulation of DNA methylation is an important epigenetic driver of AML, where the hypoxic bone marrow microenvironment can help facilitate leukaemogenesis. Thus, interactions between epigenetic regulation and hypoxia signalling will have important implications for AML development and treatment. Main body: This review summarises the importance of DNA methylation and the hypoxic bone marrow microenvironment in the development, progression, and treatment of AML. Here, we focus on the role hypoxia plays on signalling and the subsequent regulation of DNA methylation. Hypoxia is likely to influence DNA methylation through altered metabolic pathways, transcriptional control of epigenetic regulators, and direct effects on the enzymatic activity of epigenetic modifiers. DNA methylation may also prevent activation of hypoxia-responsive genes, demonstrating bidirectional crosstalk between epigenetic regulation and the hypoxic microenvironment. Finally, we consider the clinical implications of these interactions, suggesting that reduced cell cycling within the hypoxic bone marrow may decrease the efficacy of hypomethylating agents. Conclusion: Hypoxia is likely to influence AML progression through complex interactions with DNA methylation, where the therapeutic efficacy of hypomethylating agents may be limited within the hypoxic bone marrow. To achieve optimal outcomes for AML patients, future studies should therefore consider co-treatments that can promote cycling of AML cells within the bone marrow or encourage their dissociation from the bone marrow.
|
Nova | |||||||||
| 2023 |
Murray HC, Miller K, Brzozowski JS, Kahl RGS, Smith ND, Humphrey SJ, et al., 'Synergistic Targeting of DNA-PK and KIT Signaling Pathways in KIT Mutant Acute Myeloid Leukemia.', Mol Cell Proteomics, 22 100503 (2023) [C1]
|
Nova | |||||||||
| 2023 |
Foster JB, Alonso MM, Sayour E, Davidson TB, Persson ML, Dun MD, et al., 'Translational considerations for immunotherapy clinical trials in pediatric neuro-oncology', Neoplasia (United States), 42 (2023) [C1] While immunotherapy for pediatric cancer has made great strides in recent decades, including the FDA approval of agents such as dinutuximab and tisgenlecleucel, these successes ha... [more] While immunotherapy for pediatric cancer has made great strides in recent decades, including the FDA approval of agents such as dinutuximab and tisgenlecleucel, these successes have rarely impacted children with pediatric central nervous system (CNS) tumors. As our understanding of the biological underpinnings of these tumors evolves, new immunotherapeutics are undergoing rapid clinical translation specifically designed for children with CNS tumors. Most recently, there have been notable clinical successes with oncolytic viruses, vaccines, adoptive cellular therapy, and immune checkpoint inhibition. In this article, the immunotherapy working group of the Pacific Pediatric Neuro-Oncology Consortium (PNOC) reviews the current and future state of immunotherapeutic CNS clinical trials with a focus on clinical trial development. Based on recent therapeutic trials, we discuss unique immunotherapy clinical trial challenges, including toxicity considerations, disease assessment, and correlative studies. Combinatorial strategies and future directions will be addressed. Through internationally collaborative efforts and consortia, we aim to direct this promising field of immuno-oncology to the next frontier of successful application against pediatric CNS tumors.
|
Nova | |||||||||
| 2023 |
Dun MD, Odia Y, Arrillaga-Romany I, 'Diffuse midline glioma, H3K27-altered: Illuminating the dark side of the moon.', Neuro Oncol, (2023) [C1]
|
||||||||||
| 2022 |
Findlay IJ, De Iuliis GN, Duchatel RJ, Jackson ER, Vitanza NA, Cain JE, et al., 'Pharmaco-proteogenomic profiling of pediatric diffuse midline glioma to inform future treatment strategies', Oncogene, 41 461-475 (2022) [C1] Diffuse midline glioma (DMG) is a deadly pediatric and adolescent central nervous system (CNS) tumor localized along the midline structures of the brain atop the spinal cord. With... [more] Diffuse midline glioma (DMG) is a deadly pediatric and adolescent central nervous system (CNS) tumor localized along the midline structures of the brain atop the spinal cord. With a median overall survival (OS) of just 9¿11-months, DMG is characterized by global hypomethylation of histone H3 at lysine 27 (H3K27me3), driven by recurring somatic mutations in H3 genes including, HIST1H3B/C (H3.1K27M) or H3F3A (H3.3K27M), or through overexpression of EZHIP in patients harboring wildtype H3. The recent World Health Organization¿s 5th Classification of CNS Tumors now designates DMG as, ¿H3 K27-altered¿, suggesting that global H3K27me3 hypomethylation is a ubiquitous feature of DMG and drives devastating transcriptional programs for which there are no treatments. H3-alterations co-segregate with various other somatic driver mutations, highlighting the high-level of intertumoral heterogeneity of DMG. Furthermore, DMG is also characterized by very high-level intratumoral diversity with tumors harboring multiple subclones within each primary tumor. Each subclone contains their own combinations of driver and passenger lesions that continually evolve, making precision-based medicine challenging to successful execute. Whilst the intertumoral heterogeneity of DMG has been extensively investigated, this is yet to translate to an increase in patient survival. Conversely, our understanding of the non-genomic factors that drive the rapid growth and fatal nature of DMG, including endogenous and exogenous microenvironmental influences, neurological cues, and the posttranscriptional and posttranslational architecture of DMG remains enigmatic or at best, immature. However, these factors are likely to play a significant role in the complex biological sequelae that drives the disease. Here we summarize the heterogeneity of DMG and emphasize how analysis of the posttranslational architecture may improve treatment paradigms. We describe factors that contribute to treatment response and disease progression, as well as highlight the potential for pharmaco-proteogenomics (i.e., the integration of genomics, proteomics and pharmacology) in the management of this uniformly fatal cancer.
|
Nova | |||||||||
| 2022 |
Przystal JM, Cosentino CC, Yadavilli S, Zhang J, Laternser S, Bonner ER, et al., 'Imipridones affect tumor bioenergetics and promote cell lineage differentiation in diffuse midline gliomas', Neuro-Oncology, 24 1438-1451 (2022) [C1]
|
Nova | |||||||||
| 2022 |
Calvert L, Green MP, De Iuliis GN, Dun MD, Turner BD, Clarke BO, et al., 'Assessment of the Emerging Threat Posed by Perfluoroalkyl and Polyfluoroalkyl Substances to Male Reproduction in Humans', FRONTIERS IN ENDOCRINOLOGY, 12 (2022) [C1]
|
Nova | |||||||||
| 2022 |
Hunt K, Burnard SM, Roper EA, Bond DR, Dun MD, Verrills NM, et al., 'scTEM-seq: Single-cell analysis of transposable element methylation to link global epigenetic heterogeneity with transcriptional programs', SCIENTIFIC REPORTS, 12 (2022) [C1]
|
Nova | |||||||||
| 2022 |
Liu I, Jiang L, Samuelsson ER, Marco Salas S, Beck A, Hack OA, et al., 'The landscape of tumor cell states and spatial organization in H3-K27M mutant diffuse midline glioma across age and location', Nature Genetics, 54 1881-1894 (2022) [C1] Histone 3 lysine27-to-methionine (H3-K27M) mutations most frequently occur in diffuse midline gliomas (DMGs) of the childhood pons but are also increasingly recognized in adults. ... [more] Histone 3 lysine27-to-methionine (H3-K27M) mutations most frequently occur in diffuse midline gliomas (DMGs) of the childhood pons but are also increasingly recognized in adults. Their potential heterogeneity at different ages and midline locations is vastly understudied. Here, through dissecting the single-cell transcriptomic, epigenomic and spatial architectures of a comprehensive cohort of patient H3-K27M DMGs, we delineate how age and anatomical location shape glioma cell-intrinsic and -extrinsic features in light of the shared driver mutation. We show that stem-like oligodendroglial precursor-like cells, present across all clinico-anatomical groups, display varying levels of maturation dependent on location. We reveal a previously underappreciated relationship between mesenchymal cancer cell states and age, linked to age-dependent differences in the immune microenvironment. Further, we resolve the spatial organization of H3-K27M DMG cell populations and identify a mitotic oligodendroglial-lineage niche. Collectively, our study provides a powerful framework for rational modeling and therapeutic interventions.
|
Nova | |||||||||
| 2022 |
Li X, Liu H, Dun MD, Faulkner S, Liu X, Jiang CC, Hondermarck H, 'Proteome and secretome analysis of pancreatic cancer cells', PROTEOMICS, 22 (2022) [C1]
|
Nova | |||||||||
| 2022 |
Skerrett-Byrne DA, Anderson AL, Bromfield EG, Bernstein IR, Mulhall JE, Schjenken JE, et al., 'Global profiling of the proteomic changes associated with the post-testicular maturation of mouse spermatozoa', Cell Reports, 41 (2022) [C1] Spermatozoa acquire fertilization potential during passage through a highly specialized region of the extratesticular ductal system known as the epididymis. In the absence of de n... [more] Spermatozoa acquire fertilization potential during passage through a highly specialized region of the extratesticular ductal system known as the epididymis. In the absence of de novo gene transcription or protein translation, this functional transformation is extrinsically driven via the exchange of varied macromolecular cargo between spermatozoa and the surrounding luminal plasma. Key among these changes is a substantive remodeling of the sperm proteomic architecture, the scale of which has yet to be fully resolved. Here, we have exploited quantitative mass spectrometry-based proteomics to define the extent of changes associated with the maturation of mouse spermatozoa; reporting the identity of >6,000 proteins, encompassing the selective loss and gain of several hundred proteins. Further, we demonstrate epididymal-driven activation of RHOA-mediated signaling pathways is an important component of sperm maturation. These data contribute molecular insights into the complexity of proteomic changes associated with epididymal sperm maturation.
|
Nova | |||||||||
| 2022 |
Persson ML, Douglas AM, Alvaro F, Faridi P, Larsen MR, Alonso MM, et al., 'The intrinsic and microenvironmental features of diffuse midline glioma: Implications for the development of effective immunotherapeutic treatment strategies', NEURO-ONCOLOGY, 24 1408-1422 (2022) [C1]
|
Nova | |||||||||
| 2022 |
Staudt DE, Murray HC, Skerrett-Byrne DA, Smith ND, Jamaluddin MFB, Kahl RGS, et al., 'Phospho-heavy-labeled-spiketide FAIMS stepped-CV DDA (pHASED) provides real-time phosphoproteomics data to aid in cancer drug selection', CLINICAL PROTEOMICS, 19 (2022) [C1]
|
Nova | |||||||||
| 2022 |
McLachlan T, Matthews WC, Jackson ER, Staudt DE, Douglas AM, Findlay IJ, et al., 'B-cell Lymphoma 6 (BCL6): From Master Regulator of Humoral Immunity to Oncogenic Driver in Pediatric Cancers', Molecular cancer research : MCR, 20 1711-1723 (2022) [C1] B-cell lymphoma 6 (BCL6) is a protooncogene in adult and pediatric cancers, first identified in diffuse large B-cell lymphoma (DLBCL) where it acts as a repressor of the tumor sup... [more] B-cell lymphoma 6 (BCL6) is a protooncogene in adult and pediatric cancers, first identified in diffuse large B-cell lymphoma (DLBCL) where it acts as a repressor of the tumor suppressor TP53, conferring survival, protection, and maintenance of lymphoma cells. BCL6 expression in normal B cells is fundamental in the regulation of humoral immunity, via initiation and maintenance of the germinal centers (GC). Its role in B cells during the production of high affinity immunoglobins (that recognize and bind specific antigens) is believed to underpin its function as an oncogene. BCL6 is known to drive the self-renewal capacity of leukemia-initiating cells (LIC), with high BCL6 expression in acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and glioblastoma (GBM) associated with disease progression and treatment resistance. The mechanisms underpinning BCL6-driven therapy resistance are yet to be uncovered; however, high activity is considered to confer poor prognosis in the clinical setting. BCL6's key binding partner, BCL6 corepressor (BCOR), is frequently mutated in pediatric cancers and appears to act in concert with BCL6. Using publicly available data, here we show that BCL6 is ubiquitously overexpressed in pediatric brain tumors, inversely to BCOR, highlighting the potential for targeting BCL6 in these often lethal and untreatable cancers. In this review, we summarize what is known of BCL6 (role, effect, mechanisms) in pediatric cancers, highlighting the two sides of BCL6 function, humoral immunity, and tumorigenesis, as well as to review BCL6 inhibitors and highlight areas of opportunity to improve the outcomes of patients with pediatric cancer.
|
Nova | |||||||||
| 2021 |
Nixon B, Anderson AL, Bromfield EG, Martin JH, Cafe SL, Skerrett-Byrne DA, et al., 'Post-testicular sperm maturation in the saltwater crocodile Crocodylus porosus: assessing the temporal acquisition of sperm motility', REPRODUCTION FERTILITY AND DEVELOPMENT, 33 530-539 (2021) [C1]
|
Nova | |||||||||
| 2021 |
Mannan A, Germon ZP, Chamberlain J, Sillar JR, Nixon B, Dun MD, 'Reactive oxygen species in acute lymphoblastic leukaemia: Reducing radicals to refine responses', Antioxidants, 10 (2021) [C1]
|
Nova | |||||||||
| 2021 |
Skerrett-Byrne DA, Bromfield EG, Murray HC, Jamaluddin MFB, Jarnicki AG, Fricker M, et al., 'Time-resolved proteomic profiling of cigarette smoke-induced experimental chronic obstructive pulmonary disease', Respirology, 26 960-973 (2021) [C1] Background and objective: Chronic obstructive pulmonary disease (COPD) is the third leading cause of illness and death worldwide. Current treatments aim to control symptoms with n... [more] Background and objective: Chronic obstructive pulmonary disease (COPD) is the third leading cause of illness and death worldwide. Current treatments aim to control symptoms with none able to reverse disease or stop its progression. We explored the major molecular changes in COPD pathogenesis. Methods: We employed quantitative label-based proteomics to map the changes in the lung tissue proteome of cigarette smoke-induced experimental COPD that is induced over 8 weeks and progresses over 12 weeks. Results: Quantification of 7324 proteins enabled the tracking of changes to the proteome. Alterations in protein expression profiles occurred in the induction phase, with 18 and 16 protein changes at 4- and 6-week time points, compared to age-matched controls, respectively. Strikingly, 269 proteins had altered expression after 8 weeks when the hallmark pathological features of human COPD emerge, but this dropped to 27 changes at 12 weeks with disease progression. Differentially expressed proteins were validated using other mouse and human COPD bronchial biopsy samples. Major changes in RNA biosynthesis (heterogeneous nuclear ribonucleoproteins C1/C2 [HNRNPC] and RNA-binding protein Musashi homologue 2 [MSI2]) and modulators of inflammatory responses (S100A1) were notable. Mitochondrial dysfunction and changes in oxidative stress proteins also occurred. Conclusion: We provide a detailed proteomic profile, identifying proteins associated with the pathogenesis and disease progression of COPD establishing a platform to develop effective new treatment strategies.
|
Nova | |||||||||
| 2021 |
Skerrett-Byrne DA, Trigg NA, Bromfield EG, Dun MD, Bernstein IR, Anderson AL, et al., 'Proteomic dissection of the impact of environmental exposures on mouse seminal vesicle function', Molecular and Cellular Proteomics, 20 (2021) [C1] Seminal vesicles are an integral part of the male reproductive accessory gland system. They produce a complex array of secretions containing bioactive constituents that support ga... [more] Seminal vesicles are an integral part of the male reproductive accessory gland system. They produce a complex array of secretions containing bioactive constituents that support gamete function and promote reproductive success, with emerging evidence suggesting these secretions are influenced by our environment. Despite their significance, the biology of seminal vesicles remains poorly defined. Here, we complete the first proteomic assessment of mouse seminal vesicles and assess the impact of the reproductive toxicant acrylamide. Mice were administered acrylamide (25 mg/kg bw/day) or control daily for five consecutive days prior to collecting seminal vesicle tissue. A total of 5013 proteins were identified in the seminal vesicle proteome with bioinformatic analyses identifying cell proliferation, protein synthesis, cellular death, and survival pathways as prominent biological processes. Secreted proteins were among the most abundant, and several proteins are linked with seminal vesicle phenotypes. Analysis of the effect of acrylamide on the seminal vesicle proteome revealed 311 differentially regulated (FC ± 1.5, p = 0.05, 205 up-regulated, 106 downregulated) proteins, orthogonally validated via immunoblotting and immunohistochemistry. Pathways that initiate protein synthesis to promote cellular survival were prominent among the dysregulated pathways, and rapamycin-insensitive companion of mTOR (RICTOR, p = 6.69E-07) was a top-ranked upstream driver. Oxidative stress was implicated as contributing to protein changes, with acrylamide causing an increase in 8-OHdG in seminal vesicle epithelial cells (fivefold increase, p = 0.016) and the surrounding smooth muscle layer (twofold increase, p = 0.043). Additionally, acrylamide treatment caused a reduction in seminal vesicle secretion weight (36% reduction, p = 0.009) and total protein content (25% reduction, p = 0.017). Together these findings support the interpretation that toxicant exposure influences male accessory gland physiology and highlights the need to consider the response of all male reproductive tract tissues when interpreting the impact of environmental stressors on male reproductive function.
|
Nova | |||||||||
| 2021 |
Skerrett-Byrne DA, Anderson AL, Hulse L, Wass C, Dun MD, Bromfield EG, et al., 'Proteomic analysis of koala (phascolarctos cinereus) spermatozoa and prostatic bodies', Proteomics, 21 (2021) [C1] The aims of this study were to investigate the proteome of koala spermatozoa and that of the prostatic bodies with which they interact during ejaculation. For this purpose, sperma... [more] The aims of this study were to investigate the proteome of koala spermatozoa and that of the prostatic bodies with which they interact during ejaculation. For this purpose, spermatozoa and prostatic bodies were fractionated from the semen of four male koalas and analysed by HPLC MS/MS. This strategy identified 744 sperm and 1297 prostatic body proteins, which were subsequently attributed to 482 and 776 unique gene products, respectively. Gene ontology curation of the sperm proteome revealed an abundance of proteins mapping to the canonical sirtuin and 14-3-3 signalling pathways. By contrast, protein ubiquitination and unfolded protein response pathways dominated the equivalent analysis of proteins uniquely identified in prostatic bodies. Koala sperm proteins featured an enrichment of those mapping to the functional categories of cellular compromise/inflammatory response, whilst those of the prostatic body revealed an over-representation of molecular chaperone and stress-related proteins. Cross-species comparisons demonstrated that the koala sperm proteome displays greater conservation with that of eutherians (human; 93%) as opposed to reptile (crocodile; 39%) and avian (rooster; 27%) spermatozoa. Together, this work contributes to our overall understanding of the core sperm proteome and has identified biomarkers that may contribute to the exceptional longevity of koala spermatozoa during ex vivo storage.
|
Nova | |||||||||
| 2021 |
Duchatel RJ, Mannan A, Woldu AS, Hawtrey T, Hindley PA, Douglas AM, et al., 'Preclinical and clinical evaluation of German-sourced ONC201 for the treatment of H3K27M-mutant diffuse intrinsic pontine glioma.', Neuro-oncology advances, 3 vdab169 (2021) [C1]
|
Nova | |||||||||
| 2021 |
Trigg NA, Skerrett-Byrne DA, Xavier MJ, Zhou W, Anderson AL, Stanger SJ, et al., 'Acrylamide modulates the mouse epididymal proteome to drive alterations in the sperm small non-coding RNA profile and dysregulate embryo development', Cell Reports, 37 (2021) [C1] Paternal exposure to environmental stressors elicits distinct changes to the sperm sncRNA profile, modifications that have significant post-fertilization consequences. Despite thi... [more] Paternal exposure to environmental stressors elicits distinct changes to the sperm sncRNA profile, modifications that have significant post-fertilization consequences. Despite this knowledge, there remains limited mechanistic understanding of how paternal exposures modify the sperm sncRNA landscape. Here, we report the acute sensitivity of the sperm sncRNA profile to the reproductive toxicant acrylamide. Furthermore, we trace the differential accumulation of acrylamide-responsive sncRNAs to coincide with sperm transit of the proximal (caput) segment of the epididymis, wherein acrylamide exposure alters the abundance of several transcription factors implicated in the expression of acrylamide-sensitive sncRNAs. We also identify extracellular vesicles secreted from the caput epithelium in relaying altered sncRNA profiles to maturing spermatozoa and dysregulated gene expression during early embryonic development following fertilization by acrylamide-exposed spermatozoa. These data provide mechanistic links to account for how environmental insults can alter the sperm epigenome and compromise the transcriptomic profile of early embryos.
|
Nova | |||||||||
| 2021 |
Ruhen O, Qu X, Jamaluddin MFB, Salomon C, Gandhi A, Millward M, et al., 'Dynamic Landscape of Extracellular Vesicle-Associated Proteins Is Related to Treatment Response of Patients with Metastatic Breast Cancer', MEMBRANES, 11 (2021) [C1]
|
Nova | |||||||||
| 2021 |
Trigg NA, Stanger SJ, Zhou W, Skerrett-Byrne DA, Sipilä P, Dun MD, et al., 'A novel role for milk fat globule-EGF factor 8 protein (MFGE8) in the mediation of mouse sperm extracellular vesicle interactions', Proteomics, 21 (2021) [C1] Spermatozoa transition to functional maturity as they are conveyed through the epididymis, a highly specialized region of the male excurrent duct system. Owing to their transcript... [more] Spermatozoa transition to functional maturity as they are conveyed through the epididymis, a highly specialized region of the male excurrent duct system. Owing to their transcriptionally and translationally inert state, this transformation into fertilization competent cells is driven by complex mechanisms of intercellular communication with the secretory epithelium that delineates the epididymal tubule. Chief among these mechanisms are the release of extracellular vesicles (EV), which have been implicated in the exchange of varied macromolecular cargo with spermatozoa. Here, we describe the optimization of a tractable cell culture model to study the mechanistic basis of sperm¿extracellular vesicle interactions. In tandem with receptor inhibition strategies, our data demonstrate the importance of milk fat globule-EGF factor 8 (MFGE8) protein in mediating the efficient exchange of macromolecular EV cargo with mouse spermatozoa; with the MFGE8 integrin-binding Arg-Gly-Asp (RGD) tripeptide motif identified as being of particular importance. Specifically, complementary strategies involving MFGE8 RGD domain ablation, competitive RGD-peptide inhibition and antibody-masking of alpha V integrin receptors, all significantly inhibited the uptake and redistribution of EV-delivered proteins into immature mouse spermatozoa. These collective data implicate the MFGE8 ligand and its cognate integrin receptor in the mediation of the EV interactions that underpin sperm maturation.
|
Nova | |||||||||
| 2020 |
Nixon B, Cafe SL, Eamens AL, De Iuliis GN, Bromfield EG, Martin JH, et al., 'Molecular insights into the divergence and diversity of post-testicular maturation strategies', Molecular and Cellular Endocrinology, 517 110955-110955 (2020) [C1]
|
Nova | |||||||||
| 2020 |
Cafe SL, Nixon B, Dun MD, Roman SD, Bernstein IR, Bromfield EG, 'Oxidative Stress Dysregulates Protein Homeostasis within the Male Germ Line', Antioxidants and Redox Signaling, 32 487-503 (2020) [C1]
|
Nova | |||||||||
| 2020 |
Liu H, Li X, Dun MD, Faulkner S, Jiang CC, Hondermarck H, 'Cold Shock Domain Containing E1 (CSDE1) Protein is Overexpressed and Can be Targeted to Inhibit Invasiveness in Pancreatic Cancer Cells', PROTEOMICS, 20 (2020) [C1]
|
Nova | |||||||||
| 2020 |
Stanger SJ, Bernstein IR, Anderson AL, Hutcheon K, Dun MD, Eamens AL, Nixon B, 'The abundance of a transfer RNA-derived RNA fragment small RNA subpopulation is enriched in cauda spermatozoa', ExRNA, 2 (2020) [C1]
|
Nova | |||||||||
| 2020 |
Afrin F, Chi M, Eamens AL, Duchatel RJ, Douglas AM, Schneider J, et al., 'Can hemp help? Low-THC cannabis and non-THC cannabinoids for the treatment of cancer', Cancers, 12 (2020) [C1]
|
Nova | |||||||||
| 2020 |
Dun MD, Mannan A, Rigby CJ, Butler S, Toop HD, Beck D, et al., 'Shwachman Bodian Diamond syndrome (SBDS) protein is a direct inhibitor of protein phosphatase 2A (PP2A) activity and overexpressed in acute myeloid leukaemia', Leukemia, 34 3393-3397 (2020) [C1]
|
Nova | |||||||||
| 2019 |
Sillar JR, Germon ZP, DeIuliis GN, Dun MD, 'The Role of Reactive Oxygen Species in Acute Myeloid Leukaemia.', International Journal of Molecular Sciences, 20 (2019) [C1]
|
Nova | |||||||||
| 2019 |
Nixon B, De Iuliis GN, Dun MD, Zhou W, Trigg NA, Eamens AL, 'Profiling of epididymal small non-protein-coding RNAs', Andrology, 7 669-680 (2019) [C1]
|
Nova | |||||||||
| 2019 |
Nixon B, De Iuliis GN, Hart HM, Zhou W, Mathe A, Bernstein IR, et al., 'Proteomic profiling of mouse epididymosomes reveals their contributions to post-testicular sperm maturation', Molecular and Cellular Proteomics, 18 S91-S108 (2019) [C1]
|
Nova | |||||||||
| 2019 |
Duchatel RJ, Jackson ER, Alvaro F, Nixon B, Hondermarck H, Dun MD, 'Signal Transduction in Diffuse Intrinsic Pontine Glioma', PROTEOMICS, 19 (2019) [C1]
|
Nova | |||||||||
| 2019 |
Nixon B, Johnston SD, Skerrett-Byrne DA, Anderson AL, Stanger SJ, Bromfield EG, et al., 'Modification of Crocodile Spermatozoa Refutes the Tenet That Post-testicular Sperm Maturation Is Restricted To Mammals', MOLECULAR & CELLULAR PROTEOMICS, 18 S59-S76 (2019) [C1]
|
Nova | |||||||||
| 2019 |
Nixon B, Bernstein IR, Cafe SL, Delehedde M, Sergeant N, Anderson AL, et al., 'A Kinase Anchor Protein 4 is vulnerable to oxidative adduction in male germ cells', Frontiers in Cell and Developmental Biology, 7 (2019) [C1]
|
Nova | |||||||||
| 2019 |
Zhou W, Stanger SJ, Anderson AL, Bernstein IR, De Iuliis GN, McCluskey A, et al., 'Mechanisms of tethering and cargo transfer during epididymosome-sperm interactions.', BMC biology, 17 35-35 (2019) [C1]
|
Nova | |||||||||
| 2019 |
Bromfield E, Walters JLH, Cafe S, Bernstein I, Stanger SR, Anderson AL, et al., 'Differential cell death decisions in the testis: evidence for an exclusive window of ferroptosis in round spermatids', Molecular Human Reproduction, 25 241-256 (2019) [C1]
|
Nova | |||||||||
| 2019 |
Petit J, Carroll G, Gould T, Pockney P, Dun M, Scott RJ, 'Cell-free DNA as a Diagnostic Blood-Based Biomarker for Colorectal Cancer: A Systematic Review', Journal of Surgical Research, 236 184-197 (2019) [C1] Background: Circulating tumour DNA (ctDNA) has emerged as an excellent candidate for the future of liquid biopsies for many cancers. There has been growing interest in blood-based... [more] Background: Circulating tumour DNA (ctDNA) has emerged as an excellent candidate for the future of liquid biopsies for many cancers. There has been growing interest in blood-based liquid biopsy because of the potential of ctDNA to produce a noninvasive test that can be used for: the diagnosis of colorectal cancer, monitoring therapy response, and providing information on overall prognosis. The aim of this review was to collate and explore the current evidence regarding ctDNA as a screening tool for colorectal cancer (CRC). Methods: A systematic review of published articles in English over the past 20 y was performed using Medline, Embase, and Cochrane databases on May 23, 2017. After a full-text review, a total of 69 studies were included. Two assessment tools were used to review and compare the methodological quality of these studies. Results: Among the 69 studies included, 17 studies reviewed total cfDNA, whereas six studies looked at the DNA integrity index and 15 focused on ctDNA. There were a total of 40 studies that reviewed methylated cfDNA with 19 of these focussing specifically on SEPT9. Conclusions: The results of this review indicate that methylated epigenetic ctDNA markers are perhaps the most promising candidates for a blood-based CRC-screening modality using cell-free (cf) DNA. Methylated cfDNA appears to be less specific for CRC compared to ctDNA; however, they have demonstrated good sensitivity for early-stage CRC. Further research is required to determine which methylated cfDNA markers are the most accurate when applied to large cohorts of patients. In addition, reliable comparison of results across multiple studies would benefit from standardization of methodology for DNA extraction and PCR techniques in the future.
|
Nova | |||||||||
| 2018 |
Zhou W, De Iuliis GN, Dun MD, Nixon B, 'Characteristics of the epididymal luminal environment responsible for sperm maturation and storage', Frontiers in Endocrinology, 9 (2018) [C1]
|
Nova | |||||||||
| 2018 |
Almazi JG, Pockney P, Gedye C, Smith ND, Hondermarck H, Verrills NM, Dun MD, 'Cell-Free DNA Blood Collection Tubes Are Appropriate for Clinical Proteomics: A Demonstration in Colorectal Cancer.', Proteomics. Clinical applications, 12 e1700121 (2018) [C1]
|
Nova | |||||||||
| 2018 |
Wang TE, Li SH, Minabe S, Andreson AL, Dun MD, Maeda KI, et al., 'Mouse quiescin sulfhydryl oxidases exhibit distinct epididymal luminal distribution with segment-specific sperm surface associations', Biology of Reproduction, 99 1022-1033 (2018) [C1]
|
Nova | |||||||||
| 2018 |
Staudt D, Murray HC, McLachlan T, Alvaro F, Enjeti AK, Verrills NM, Dun MD, 'Targeting Oncogenic Signaling in Mutant FLT3 Acute Myeloid Leukemia: The Path to Least Resistance', INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES, 19 (2018) [C1]
|
Nova | |||||||||
| 2018 |
Zhou W, Sipilä P, De Iuliis G, Dun MD, Nixon B, 'Analysis of Epididymal Protein Synthesis and Secretion', Jove-Journal of Visualized Experiments, 138 (2018) [C1]
|
Nova | |||||||||
| 2018 |
Brzozowski JS, Bond DR, Jankowski H, Goldie BJ, Burchell R, Naudin C, et al., 'Extracellular vesicles with altered tetraspanin CD9 and CD151 levels confer increased prostate cell motility and invasion', Scientific Reports, 8 1-13 (2018) [C1]
|
Nova | |||||||||
| 2018 |
Li X, Dun MD, Faulkner S, Hondermarck H, 'Neuroproteins in Cancer: Assumed Bystanders Become Culprits', PROTEOMICS, 18 (2018) [C1]
|
Nova | |||||||||
| 2018 |
Walters JLH, De Iuliis GN, Dun MD, Aitken RJ, McLaughlin EA, Nixon B, Bromfield EG, 'Pharmacological inhibition of arachidonate 15-lipoxygenase protects human spermatozoa against oxidative stress.', Biology of reproduction, 98 784-794 (2018) [C1]
|
Nova | |||||||||
| 2018 |
Jamaluddin MFB, Ko YA, Kumar M, Brown Y, Bajwa P, Nagendra PB, et al., 'Proteomic profiling of human uterine fibroids reveals upregulation of the extracellular matrix protein periostin', Endocrinology, 159 1106-1118 (2018) [C1] The central characteristic of uterine fibroids is excessive deposition of extracellular matrix (ECM), which contributes to fibroid growth and bulk-type symptoms. Despite this, ver... [more] The central characteristic of uterine fibroids is excessive deposition of extracellular matrix (ECM), which contributes to fibroid growth and bulk-type symptoms. Despite this, very little is known about patterns of ECM protein expression in fibroids and whether these are influenced by the most common genetic anomalies, which relate to MED12. We performed extensive genetic and proteomic analyses of clinically annotated fibroids and adjacent normal myometrium to identify the composition and expression patterns of ECM proteins in MED12 mutation-positive and mutation-negative uterine fibroids. Genetic sequencing of tissue samples revealed MED12 alterations in 39 of 65 fibroids (60%) from 14 patients. Using isobaric tagged-based quantitative mass spectrometry on three selected patients (n = 9 fibroids), we observed a common set of upregulated (.1.5-fold) and downregulated (,0.66-fold) proteins in small, medium, and large fibroid samples of annotated MED12 status. These two sets of upregulated and downregulated proteins were the same in all patients, regardless of variations in fibroid size and MED12 status. We then focused on one of the significant upregulated ECM proteins and confirmed the differential expression of periostin using western blotting and immunohistochemical analysis. Our study defined the proteome of uterine fibroids and identified that increased ECM protein expression, in particular periostin, is a hallmark of uterine fibroids regardless of MED12 mutation status. This study sets the foundation for further investigations to analyze the mechanisms regulating ECM overexpression and the functional role of upregulated ECM proteins in leiomyogenesis.
|
Nova | |||||||||
| 2018 |
Degryse S, De Bock CE, Demeyer S, Govaerts I, Bornschein S, Verbeke D, et al., 'Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia', Leukemia, 32 788-800 (2018) [C1] Mutations in the interleukin-7 receptor (IL7R) or the Janus kinase 3 (JAK3) kinase occur frequently in T-cell acute lymphoblastic leukemia (T-ALL) and both are able to drive cellu... [more] Mutations in the interleukin-7 receptor (IL7R) or the Janus kinase 3 (JAK3) kinase occur frequently in T-cell acute lymphoblastic leukemia (T-ALL) and both are able to drive cellular transformation and the development of T-ALL in mouse models. However, the signal transduction pathways downstream of JAK3 mutations remain poorly characterized. Here we describe the phosphoproteome downstream of the JAK3(L857Q)/(M511I) activating mutations in transformed Ba/F3 lymphocyte cells. Signaling pathways regulated by JAK3 mutants were assessed following acute inhibition of JAK1/JAK3 using the JAK kinase inhibitors ruxolitinib or tofacitinib. Comprehensive network interrogation using the phosphoproteomic signatures identified significant changes in pathways regulating cell cycle, translation initiation, mitogen-activated protein kinase and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT signaling, RNA metabolism, as well as epigenetic and apoptotic processes. Key regulatory proteins within pathways that showed altered phosphorylation following JAK inhibition were targeted using selumetinib and trametinib (MEK), buparlisib (PI3K) and ABT-199 (BCL2), and found to be synergistic in combination with JAK kinase inhibitors in primary T-ALL samples harboring JAK3 mutations. These data provide the first detailed molecular characterization of the downstream signaling pathways regulated by JAK3 mutations and provide further understanding into the oncogenic processes regulated by constitutive kinase activation aiding in the development of improved combinatorial treatment regimens.
|
Nova | |||||||||
| 2017 |
Ong LK, Page S, Briggs GD, Guan L, Dun MD, Verrills NM, et al., 'Peripheral Lipopolysaccharide Challenge Induces Long-Term Changes in Tyrosine Hydroxylase Regulation in the Adrenal Medulla', Journal of Cellular Biochemistry, 118 2096-2107 (2017) [C1] Immune activation can alter the activity of adrenal chromaffin cells. The effect of immune activation by lipopolysaccharide (LPS) on the regulation of tyrosine hydroxylase (TH) in... [more] Immune activation can alter the activity of adrenal chromaffin cells. The effect of immune activation by lipopolysaccharide (LPS) on the regulation of tyrosine hydroxylase (TH) in the adrenal medulla in vivo was determined between 1 day and 6 months after LPS injection. The plasma levels of eleven cytokines were reduced 1 day after LPS injection, whereas the level for interleukin-10 was increased. The levels of all cytokines remained at control levels until 6 months when the levels of interleukin-6 and -4 were increased. One day after LPS injection, there was a decrease in TH-specific activity that may be due to decreased phosphorylation of serine 31 and 40. This decreased phosphorylation of serine 31 and 40 may be due to an increased activation of the protein phosphatase PP2A. One week after LPS injection, there was increased TH protein and increased phosphorylation of serine 40 that this was not accompanied by an increase in TH-specific activity. All TH parameters measured returned to basal levels between 1 month and 3 months. Six months after injection there was an increase in TH protein. This was associated with increased levels of the extracellular regulated kinase isoforms 1 and 2. This work shows that a single inflammatory event has the capacity to generate both short-term and long-term changes in TH regulation in the adrenal medulla of the adult animal. J. Cell. Biochem. 118: 2096¿2107, 2017. © 2016 Wiley Periodicals, Inc.
|
Nova | |||||||||
| 2017 |
Murray HC, Dun MD, Verrills NM, 'Harnessing the power of proteomics for identification of oncogenic, druggable signalling pathways in cancer', Expert Opinion on Drug Discovery, 12 431-447 (2017) [C1] Introduction: Genomic and transcriptomic profiling of tumours has revolutionised our understanding of cancer. However, the majority of tumours possess multiple mutations, and dete... [more] Introduction: Genomic and transcriptomic profiling of tumours has revolutionised our understanding of cancer. However, the majority of tumours possess multiple mutations, and determining which oncogene, or even which pathway, to target is difficult. Proteomics is emerging as a powerful approach to identify the functionally important pathways driving these cancers, and how they can be targeted therapeutically. Areas covered: The authors provide a technical overview of mass spectrometry based approaches for proteomic profiling, and review the current and emerging strategies available for the identification of dysregulated networks, pathways, and drug targets in cancer cells, with a key focus on the ability to profile cancer kinomes. The potential applications of mass spectrometry in the clinic are also highlighted. Expert opinion: The addition of proteomic information to genomic platforms¿¿proteogenomics¿¿is providing unparalleled insight in cancer cell biology. Application of improved mass spectrometry technology and methodology, in particular the ability to analyse post-translational modifications (the PTMome), is providing a more complete picture of the dysregulated networks in cancer, and uncovering novel therapeutic targets. While the application of proteomics to discovery research will continue to rise, improved workflow standardisation and reproducibility is required before mass spectrometry can enter routine clinical use.
|
Nova | |||||||||
| 2017 |
Bromfield EG, Mihalas BP, Dun MD, Aitken RJ, McLaughlin EA, Walters JLH, Nixon B, 'Inhibition of arachidonate 15-lipoxygenase prevents 4-hydroxynonenal-induced proteindamage in male germ cells', Biology of Reproduction, 96 598-609 (2017) [C1]
|
Nova | |||||||||
| 2017 |
Hutcheon K, McLaughlin EA, Stanger SJ, Bernstein IR, Dun MD, Eamens AL, Nixon B, 'Analysis of the small non-protein-coding RNA profile of mouse spermatozoa reveals specific enrichment of piRNAs within mature spermatozoa', RNA Biology, 14 1776-1790 (2017) [C1] Post-testicular sperm maturation and storage within the epididymis is a key determinant of gamete quality and fertilization competence. Here we demonstrate that mouse spermatozoa ... [more] Post-testicular sperm maturation and storage within the epididymis is a key determinant of gamete quality and fertilization competence. Here we demonstrate that mouse spermatozoa possess a complex small non-protein-coding RNA (sRNA) profile, the composition of which is markedly influenced by their epididymal transit. Thus, although microRNAs (miRNAs) are highly represented in the spermatozoa of the proximal epididymis, this sRNA class is largely diminished in mature spermatozoa of the distal epididymis. Coincident with this, a substantial enrichment in Piwi-interacting RNA (piRNA) abundance in cauda spermatozoa was detected. Further, features of cauda piRNAs, including; predominantly 29¿31 nts in length; preference for uracil at their 5' terminus; no adenine enrichment at piRNA nt 10, and; predominantly mapping to intergenic regions of the mouse genome, indicate that these piRNAs are generated by the PIWIL1-directed primary piRNA production pathway. Accordingly, PIWIL1 was detected via immunoblotting and mass spectrometry in epididymal spermatozoa. These data provide insight into the complexity and dynamic nature of the sRNA profile of spermatozoa and raise the intriguing prospect that piRNAs are generated in situ in maturing spermatozoa. Such information is of particular interest in view of the potential role for paternal sRNAs in influencing conception, embryo development and intergenerational inheritance.
|
Nova | |||||||||
| 2017 |
Watt LF, Panicker N, Mannan A, Copeland B, Kahl RGS, Dun MD, et al., 'Functional importance of PP2A regulatory subunit loss in breast cancer', Breast Cancer Research and Treatment, 166 117-131 (2017) [C1] Purpose: Protein phosphatase 2A (PP2A) is a family of serine/threonine phosphatases that regulate multiple cellular signalling pathways involved in proliferation, survival and apo... [more] Purpose: Protein phosphatase 2A (PP2A) is a family of serine/threonine phosphatases that regulate multiple cellular signalling pathways involved in proliferation, survival and apoptosis. PP2A inhibition occurs in many cancers and is considered a tumour suppressor. Deletion/downregulation of PP2A genes has been observed in breast tumours, but the functional role of PP2A subunit loss in breast cancer has not been investigated. Methods: PP2A subunit expression was examined by immunohistochemistry in human breast tumours, and by qPCR and immunoblotting in breast cancer cell lines. PP2A subunits were inhibited by shRNA, and mutant PP2A genes overexpressed, in MCF10A and MCF7 cells, and growth and signalling in standard and three-dimensional cultures were assessed. Results: Expression of PP2A-Aa, PP2A-Ba and PP2A-B'a subunits was significantly lower in primary human breast tumours and lymph node metastases, compared to normal mammary tissue. PP2A-Aa and the regulatory subunits PP2A-Ba, -Bd and -B'¿ were also reduced in breast cancer cell lines compared to normal mammary epithelial cells. Functionally, shRNA-mediated knockdown of PP2A-Ba, -B'a and -B'¿, but not PP2A-Aa, induced hyper-proliferation and large multilobular acini in MCF10A 3D cultures, characterised by activation of ERK. Expression of a breast cancer-associated PP2A-A mutant, PP2A-Aa-E64G, which inhibits binding of regulatory subunits to the PP2A core, induced a similar hyper-proliferative phenotype. Knockdown of PP2A-Ba also induced hyper-proliferation in MCF7 breast cancer cells. Conclusion: These results suggest that loss of specific PP2A regulatory subunits is functionally important in breast tumourigenesis, and support strategies to enhance PP2A activity as a therapeutic approach in breast cancer.
|
Nova | |||||||||
| 2017 |
Naudin C, Smith B, Bond DR, Dun MD, Scott RJ, Ashman LK, et al., 'Characterization of the early molecular changes in the glomeruli of Cd151 -/- mice highlights induction of mindin and MMP-10.', Scientific Reports, 7 15987-15987 (2017) [C1]
|
Nova | |||||||||
| 2017 |
de Bock CE, Hughes MR, Snyder K, Alley S, Sadeqzadeh E, Dun MD, et al., 'Protein interaction screening identifies SH3RF1 as a new regulator of FAT1 protein levels', FEBS LETTERS, 591 667-678 (2017) [C1]
|
Nova | |||||||||
| 2016 |
Toop HD, Dun MD, Ross BK, Flanagan HM, Verrills NM, Morris JC, 'Development of novel PP2A activators for use in the treatment of acute myeloid leukaemia', Organic and Biomolecular Chemistry, 14 4605-4616 (2016) [C1] AAL(S), the chiral deoxy analog of the FDA approved drug FTY720, has been shown to inhibit proliferation and apoptosis in several cancer cell lines. It has been suggested that it ... [more] AAL(S), the chiral deoxy analog of the FDA approved drug FTY720, has been shown to inhibit proliferation and apoptosis in several cancer cell lines. It has been suggested that it does this by activating protein phosphatase 2A (PP2A). Here we report the synthesis of new cytotoxic analogs of AAL(S) and the evaluation of their cytotoxicity in two myeloid cell lines, one of which is sensitive to PP2A activation. We show that these analogs activate PP2A in these cells supporting the suggested mechanism for their cytotoxic properties. Our findings identify key structural motifs required for anti-cancer effects.
|
Nova | |||||||||
| 2016 |
Smith AM, Dun MD, Lee EM, Harrison C, Kahl R, Flanagan H, et al., 'Activation of protein phosphatase 2A in FLT3+ acute myeloid leukemia cells enhances the cytotoxicity of FLT3 tyrosine kinase inhibitors', Oncotarget, 7 47465-47478 (2016) [C1] Constitutive activation of the receptor tyrosine kinase Fms-like tyrosine kinase 3 (FLT3), via co-expression of its ligand or by genetic mutation, is common in acute myeloid leuke... [more] Constitutive activation of the receptor tyrosine kinase Fms-like tyrosine kinase 3 (FLT3), via co-expression of its ligand or by genetic mutation, is common in acute myeloid leukemia (AML). In this study we show that FLT3 activation inhibits the activity of the tumor suppressor, protein phosphatase 2A (PP2A). Using BaF3 cells transduced with wildtype or mutant FLT3, we show that FLT3-induced PP2A inhibition sensitizes cells to the pharmacological PP2A activators, FTY720 and AAL(S). FTY720 and AAL(S) induced cell death and inhibited colony formation of FLT3 activated cells. Furthermore, PP2A activators reduced the phosphorylation of ERK and AKT, downstream targets shared by both FLT3 and PP2A, in FLT3/ITD+ BaF3 and MV4-11 cell lines. PP2A activity was lower in primary human bone marrow derived AML blasts compared to normal bone marrow, with blasts from FLT3-ITD patients displaying lower PP2A activity than WT-FLT3 blasts. Reduced PP2A activity was associated with hyperphosphorylation of the PP2A catalytic subunit, and reduced expression of PP2A structural and regulatory subunits. AML patient blasts were also sensitive to cell death induced by FTY720 and AAL(S), but these compounds had minimal effect on normal CD34+ bone marrow derived monocytes. Finally, PP2A activating compounds displayed synergistic effects when used in combination with tyrosine kinase inhibitors in FLT3-ITD+ cells. A combination of Sorafenib and FTY720 was also synergistic in the presence of a protective stromal microenvironment. Thus combining a PP2A activating compound and a FLT3 inhibitor may be a novel therapeutic approach for treating AML.
|
Nova | |||||||||
| 2015 |
Dun MD, Chalkley RJ, Faulkner S, Keene S, Avery-Kiejda KA, Scott RJ, et al., 'Proteotranscriptomic profiling of 231-BR breast cancer cells: Identification of potential biomarkers and therapeutic targets for brain metastasis', Molecular and Cellular Proteomics, 14 2316-2330 (2015) [C1] Brain metastases are a devastating consequence of cancer and currently there are no specific biomarkers or therapeutic targets for risk prediction, diagnosis, and treatment. Here ... [more] Brain metastases are a devastating consequence of cancer and currently there are no specific biomarkers or therapeutic targets for risk prediction, diagnosis, and treatment. Here the proteome of the brain metastatic breast cancer cell line 231-BR has been compared with that of the parental cell line MDA-MB-231, which is also metastatic but has no organ selectivity. Using SILAC and nanoLC-MS/MS, 1957 proteins were identified in reciprocal labeling experiments and 1584 were quantified in the two cell lines. A total of 152 proteins were confidently determined to be up- or down-regulated by more than twofold in 231-BR. Of note, 112/152 proteins were decreased as compared with only 40/152 that were increased, suggesting that down-regulation of specific proteins is an important part of the mechanism underlying the ability of breast cancer cells to metastasize to the brain. When matched against transcriptomic data, 43% of individual protein changes were associated with corresponding changes in mRNA, indicating that the transcript level is a limited predictor of protein level. In addition, differential miRNA analyses showed that most miRNA changes in 231-BR were up- (36/45) as compared with down-regulations (9/45). Pathway analysis revealed that proteome changes were mostly related to cell signaling and cell cycle, metabolism and extracellular matrix remodeling. The major protein changes in 231-BR were confirmed by parallel reaction monitoring mass spectrometry and consisted in increases (by more than fivefold) in the matrix metalloproteinase-1, ephrin-B1, stomatin, myc target-1, and decreases (by more than 10-fold) in transglutaminase-2, the S100 calcium-binding protein A4, and L-plastin. The clinicopathological significance of these major proteomic changes to predict the occurrence of brain metastases, and their potential value as therapeutic targets, warrants further investigation.
|
Nova | |||||||||
| 2015 |
Faulkner S, Dun MD, Hondermarck H, 'Proteogenomics: Emergence and promise', Cellular and Molecular Life Sciences, 72 953-957 (2015) [C1] Proteogenomics, or the integration of proteomics with genomics and transcriptomics, is emerging as the next step towards a unified understanding of cellular functions. Looking glo... [more] Proteogenomics, or the integration of proteomics with genomics and transcriptomics, is emerging as the next step towards a unified understanding of cellular functions. Looking globally and simultaneously at gene structure, RNA expression, protein synthesis and posttranslational modifications have become technically feasible and offer a new perspective to molecular processes. Recent publications have highlighted the value of proteogenomics in oncology for defining the molecular signature of human tumors, and translation to other areas of biomedicine and life sciences is anticipated. This minireview will discuss recent developments, challenges and perspectives in proteogenomics.
|
Nova | |||||||||
| 2015 |
Nixon B, Stanger SJ, Mihalas BP, Reilly JN, Anderson AL, Dun MD, et al., 'Next generation sequencing analysis reveals segmental patterns of microRNA expression in mouse epididymal epithelial cells', PLoS ONE, 10 (2015) [C1] The functional maturation of mammalian spermatozoa is accomplished as the cells descend through the highly specialized microenvironment of the epididymis. This dynamic environment... [more] The functional maturation of mammalian spermatozoa is accomplished as the cells descend through the highly specialized microenvironment of the epididymis. This dynamic environment is, in turn, created by the combined secretory and absorptive activity of the surrounding epithelium and displays an extraordinary level of regionalization. Although the regulatory network responsible for spatial coordination of epididymal function remains unclear, recent evidence has highlighted a novel role for the RNA interference pathway. Indeed, as noncanonical regulators of gene expression, small noncoding RNAs have emerged as key elements of the circuitry involved in regulating epididymal function and hence sperm maturation. Herein we have employed next generation sequencing technology to profile the genome-wide miRNA signatures of mouse epididymal cells and characterize segmental patterns of expression. An impressive profile of some 370 miRNAs were detected in the mouse epididymis, with a subset of these specifically identified within the epithelial cells that line the tubule (218). A majority of the latter miRNAs (75%) were detected at equivalent levels along the entire length of the mouse epididymis. We did however identify a small cohort of miRNAs that displayed highly regionalized patterns of expression, including miR-204-5p and miR-196b-5p, which were down- and up-regulated by approximately 39- and 45-fold between the caput/caudal regions, respectively. In addition we identified 79 miRNAs (representing ~ 21% of all miRNAs) as displaying conserved expression within all regions of the mouse, rat and human epididymal tissue. These included 8/14 members of let-7 family of miRNAs that have been widely implicated in the control of androgen signaling and the repression of cell proliferation and oncogenic pathways. Overall these data provide novel insights into the sophistication of the miRNA network that regulates the function of the male reproductive tract.
|
Nova | |||||||||
| 2015 |
Nixon B, Bromfield EG, Dun MD, Redgrove KA, McLaughlin EA, Aitken RJ, 'The role of the molecular chaperone heat shock protein A2 (HSPA2) in regulating human sperm-egg recognition', ASIAN JOURNAL OF ANDROLOGY, 17 568-573 (2015) [C1]
|
Nova | |||||||||
| 2014 |
Goldie BJ, Dun MD, Lin M, Smith ND, Verrills NM, Dayas CV, Cairns MJ, 'Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons.', Nucleic Acids Research, 42 9195-9208 (2014) [C1]
|
Nova | |||||||||
| 2014 |
Sadeqzadeh E, De Bock CE, Wojtalewicz N, Holt JE, Smith ND, Dun MD, et al., 'Furin processing dictates ectodomain shedding of human FAT1 cadherin', Experimental Cell Research, 323 41-55 (2014) [C1] Fat1 is a single pass transmembrane protein and the largest member of the cadherin superfamily. Mouse knockout models and in vitro studies have suggested that Fat1 influences cell... [more] Fat1 is a single pass transmembrane protein and the largest member of the cadherin superfamily. Mouse knockout models and in vitro studies have suggested that Fat1 influences cell polarity and motility. Fat1 is also an upstream regulator of the Hippo pathway, at least in lower vertebrates, and hence may play a role in growth control. In previous work we have established that FAT1 cadherin is initially cleaved by proprotein convertases to form a noncovalently linked heterodimer prior to expression on the cell surface. Such processing was not a requirement for cell surface expression, since melanoma cells expressed both unprocessed FAT1 and the heterodimer on the cell surface. Here we further establish that the site 1 (S1) cleavage step to promote FAT1 heterodimerisation is catalysed by furin and we identify the cleavage site utilised. For a number of other transmembrane receptors that undergo heterodimerisation the S1 processing step is thought to occur constitutively but the functional significance of heterodimerisation has been controversial. It has also been generally unclear as to the significance of receptor heterodimerisation with respect to subsequent post-translational proteolysis that often occurs in transmembrane proteins. Exploiting the partial deficiency of FAT1 processing in melanoma cells together with furin-deficient LoVo cells, we manipulated furin expression to demonstrate that only the heterodimer form of FAT1 is subject to cleavage and subsequent release of the extracellular domain. This work establishes S1-processing as a clear functional prerequisite for ectodomain shedding of FAT1 with general implications for the shedding of other transmembrane receptors. © 2014.
|
Nova | |||||||||
| 2014 |
Hatchwell L, Girkin J, Morten M, Collison A, Mattes J, Foster PS, et al., 'Salmeterol attenuates chemotactic responses in rhinovirus-induced exacerbation of allergic airways disease by modulating protein phosphatase 2A', Journal of Allergy and Clinical Immunology, (2014) [C1] Background: ß-Agonists are used for relief and control of asthma symptoms by reversing bronchoconstriction. They might also have anti-inflammatory properties, but the underpinning... [more] Background: ß-Agonists are used for relief and control of asthma symptoms by reversing bronchoconstriction. They might also have anti-inflammatory properties, but the underpinning mechanisms remain poorly understood. Recently, a direct interaction between formoterol and protein phosphatase 2A (PP2A) has been described in¿vitro. Objective: We sought to elucidate the molecular mechanisms by which ß-agonists exert anti-inflammatory effects in allergen-driven and rhinovirus 1B-exacerbated allergic airways disease (AAD). Methods: Mice were sensitized and then challenged with house dust mite to induce AAD while receiving treatment with salmeterol, formoterol, or salbutamol. Mice were also infected with rhinovirus 1B to exacerbate lung inflammation and therapeutically administered salmeterol, dexamethasone, or the PP2A-activating drug (S)-2-amino-4-(4-[heptyloxy]phenyl)-2-methylbutan-1-ol (AAL[S]). Results: Systemic or intranasal administration of salmeterol protected against the development of allergen- and rhinovirus-induced airway hyperreactivity and decreased eosinophil recruitment to the lungs as effectively as dexamethasone. Formoterol and salbutamol also showed anti-inflammatory properties. Salmeterol, but not dexamethasone, increased PP2A activity, which reduced CCL11, CCL20, and CXCL2 expression and reduced levels of phosphorylated extracellular signal-regulated kinase 1 and active nuclear factor ¿B subunits in the lungs. The anti-inflammatory effect of salmeterol was blocked by targeting the catalytic subunit of PP2A with small RNA interference. Conversely, increasing PP2A activity with AAL(S) abolished rhinovirus-induced airway hyperreactivity, eosinophil influx, and CCL11, CCL20, and CXCL2 expression. Salmeterol also directly activated immunoprecipitated PP2A in¿vitro isolated from human airway epithelial cells. Conclusions: Salmeterol exerts anti-inflammatory effects by increasing PP2A activity in AAD and rhinovirus-induced lung inflammation, which might potentially account for some of its clinical benefits. © 2013 American Academy of Allergy, Asthma & Immunology.
|
Nova | |||||||||
| 2013 |
Smith TB, Dun MD, Smith ND, Curry BJ, Connaughton HS, Aitken RJ, 'The presence of a truncated base excision repair pathway in human spermatozoa that is mediated by OGG1', JOURNAL OF CELL SCIENCE, 126 1488-1497 (2013) [C1]
|
Nova | |||||||||
| 2012 |
Dun MD, Anderson AL, Bromfield EG, Asquith KL, Emmett BJ, McLaughlin EA, et al., 'Investigation of the expression and functional significance of the novel mouse sperm protein, a disintegrin and metalloprotease with thrombospondin type 1 motifs number 10 (ADAMTS10)', International Journal of Andrology, 35 572-589 (2012) [C1]
|
Nova | |||||||||
| 2012 |
Dun MD, Aitken RJ, Nixon B, 'The role of molecular chaperones in spermatogenesis and the post-testicular maturation of mammalian spermatozoa', Human Reproduction Update, 18 420-435 (2012) [C1]
|
Nova | |||||||||
| 2011 |
Redgrove KA, Anderson AL, Dun MD, McLaughlin EA, O'Bryan MK, Aitken RJ, Nixon B, 'Involvement of multimeric protein complexes in mediating the capacitation-dependent binding of human spermatozoa to homologous zonae pellucidae', Developmental Biology, 356 460-474 (2011) [C1]
|
Nova | |||||||||
| 2011 |
Dun MD, Smith ND, Baker MA, Lin M, Aitken RJ, Nixon B, 'The chaperonin containing TCP1 complex (CCT/TRiC) is involved in mediating sperm-oocyte interaction', Journal of Biological Chemistry, 286 36875-36887 (2011) [C1]
|
Nova | |||||||||
| Show 89 more journal articles | |||||||||||
Conference (80 outputs)
| Year | Citation | Altmetrics | Link | |||||
|---|---|---|---|---|---|---|---|---|
| 2023 | Liu I, Jiang L, Samuelsson E, Salas SM, Beck A, Hack O, et al., 'The landscape of tumor cell states and spatial organization in H3-K27M mutant diffuse midline glioma across age and location', BRAIN PATHOLOGY, GERMANY, Berlin (2023) | |||||||
| 2023 |
Mabotuwana NS, Skerrett-Byrne DA, Ashour DE, Butel-Simoes L, Mcgee M, Rech L, et al., 'Harnessing novel paracrine factors from the proteomes of cardiac and mesenchymal stem cells to promote repair of the fibrotic heart', EUROPEAN JOURNAL OF HEART FAILURE (2023)
|
|||||||
| 2023 |
Liu I, Jiang L, Samuelsson E, Salas SM, Beck A, Hack O, et al., 'THE LANDSCAPE OF TUMOR CELL STATES AND SPATIAL ORGANIZATION IN H3-K27M MUTANT DIFFUSE MIDLINE GLIOMA ACROSS AGE AND LOCATION', NEURO-ONCOLOGY (2023)
|
|||||||
| 2023 |
Duchatel R, Jackson E, Parackal S, Sun C, Daniel P, Mannan A, et al., 'EXPLOITING THE GENETIC DEPENDENCY ON PI3K/ MTOR SIGNALING FOR THE TREATMENT OF H3-ALTERED DIFFUSE MIDLINE GLIOMA', NEURO-ONCOLOGY, CANADA, Vancouver (2023)
|
|||||||
| 2023 |
Jackson E, Duchatel R, Staudt D, Persson M, Mannan A, Yadavilli S, et al., 'COMBINING ONC201 AND PAXALISIB FOR THE TREATMENT OF DIFFUSE MIDLINE GLIOMA (DMG); THE PRECLINICAL RESULTS UNDERPINNING THE INTERNATIONAL PHASE II CLINICAL TRIAL (NCT05009992).', NEURO-ONCOLOGY, CANADA, Vancouver (2023)
|
|||||||
| 2015 |
Watt LF, Panicker N, Copeland B, Kahl RGS, Dun MD, Young B, et al., 'PP2A a novel biomarker and therapeutic target for poor outcome breast cancer', Proceedings of the Lowy Cancer Conference, Sydney (2015) [E3]
|
|||||||
| 2015 |
Dun M, Murray H, Al-mazi J, Kahl R, Flanagan H, Smith N, et al., 'IDENTIFICATION AND SYNERGISTIC TARGETING OF FLT3-ACTIVATED PATHWAYS IN ACUTE MYELOID LEUKAEMIA', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2015 |
Al-mazi J, Dun M, Smith N, Verrills N, 'DEVELOPMENT OF NOVEL MULTIPLE REACTION MONITORING (MRM) ASSAY FOR BIOMARKER QUANTITATION IN CANCER CELLS', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2015 |
Brzozowski J, Coldie B, Jankowski H, Bond D, Scarlett C, Dun M, et al., 'THE EFFECTS OF ALTERED CD9 AND CD151 EXPRESSION ON PROSTATE EXOSOMES', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2015 |
Lehman W, Kahl R, Flanagan H, Verrills N, Dun M, 'DETERMINING THE MECHANISM OF LEUKAEMOGENESIS INDUCED BY SHWACHMAN-DIAMOND SYNDROME (SDS) USING COMPARATIVE AND QUANTITATIVE PROTEOMICS', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2015 |
Li X, Flanagan H, Kahl R, Rigby C, Verrills N, Dun M, 'CHEMICAL PROTEOMICS TO IDENTIFY THE MECHANISM OF PROTEIN PHOSPHATASE 2A (PP2A) INHIBITION IN ACUTE MYELOID LEUKAEMIA (AML)', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2015 |
Mannan A, Panicker N, Watt L, Kahl R, Dun M, Skelding K, Verrills N, 'ROLE OF REDUCED PROTEIN PHOSPHATASE 2A SUBUNIT, B55A, EXPRESSION IN LUMINAL B BREAST CANCER CELL LINE DNA DAMAGE REPAIR PATHWAY', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2015 |
Panicker N, Watt L, Kahl R, Dun M, Greer P, Skelding K, Verrills N, 'REDUCED EXPRESSION OF PROTEIN PHOSPHATASE 2A SUBUNIT, B55A, IN BREAST CANCER DNA DAMAGE REPAIR PATHWAYS', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2015 |
Rigby C, Kahl R, Flanagan H, Li X, Enjeti A, Verrills N, Dun M, 'CHARACTERISATION OF A NOVEL PP2A INHIBITORY ONCOPROTEIN IN ACUTE MYELOID LEUKAEMIA (AML)', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2015 |
Udeh R, Kahl R, Flanagan H, Verrills N, Dun M, 'IDENTIFYING THE FUNCTIONAL ROLE OF TSR, A NOVEL DRUG TARGET IN AML', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2015) [E3]
|
|||||||
| 2014 |
Dun MD, Chalkley RJ, Keene S, Bradshaw RA, Hondermarck H, 'Proteomics versus Transcriptomics for the Identification of Cancer Biomarkers: the Case of Brain-derived Metastatic Breast Cancer Cells', MOLECULAR & CELLULAR PROTEOMICS (2014) [E3]
|
|||||||
| 2014 |
Dun MD, Kahl RGS, Flanagan H, Cairns MMJ, Smith ND, Enjeti AK, et al., 'IDENTIFICATION OF ONCOGENIC SIGNALLING PATHWAYS IN ACUTE MYELOID LEUKAEMIA (AML) PATIENTS BY PHOSPHOPROTEOMICS', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2014) [E3]
|
|||||||
| 2014 |
De Iuliis GN, Verrills NM, Dun MD, 'IN SILICO ANALYSIS OF THE TARGETS OF SMALL-MOLECULE, ANTI-CANCER COMPOUNDS FOR IMPROVED CANCER THERAPEUTICS', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2014) [E3]
|
|||||||
| 2014 |
Gilbert J, Dun MD, De Iuliis GN, McCluskey A, Sakoff JA, 'SELECTIVELY TARGETING BREAST CANCER CELLS VIA CHECKPOINT ACTIVATION', ASIA-PACIFIC JOURNAL OF CLINICAL ONCOLOGY (2014) [E3]
|
|||||||
| 2013 |
Dun MD, Smith AM, Kahl RGS, Smith ND, Khanna A, Don AS, et al., 'Unraveling the mechanism of action: drugs that activate the tumor suppressor 2A.', CANCER RESEARCH, Washington, DC (2013) [E3]
|
|||||||
| 2010 |
Dun MD, Aitken RJ, Nixon B, 'The Chaperonin Containing TCP-1 (CCT/TRIC) Multisubunit Complex is Involved in Mediating Sperm-Oocyte Interactions', Reproduction, Fertility and Development, Sydney (2010) [E3]
|
|||||||
| Show 77 more conferences | ||||||||
Other (5 outputs)
| Year | Citation | Altmetrics | Link | |||||
|---|---|---|---|---|---|---|---|---|
| 2023 |
Jackson ER, Duchatel RJ, Staudt DE, Persson ML, Mannan A, Yadavilli S, et al., 'Supplementary Data from ONC201 in Combination with Paxalisib for the Treatment of H3K27-Altered Diffuse Midline Glioma', American Association for Cancer Research (AACR) (2023)
|
|||||||
| 2023 |
Jackson ER, Duchatel RJ, Staudt DE, Persson ML, Mannan A, Yadavilli S, et al., 'Supplementary Data from ONC201 in Combination with Paxalisib for the Treatment of H3K27-Altered Diffuse Midline Glioma', American Association for Cancer Research (AACR) (2023)
|
|||||||
| 2023 |
Jackson ER, Duchatel RJ, Staudt DE, Persson ML, Mannan A, Yadavilli S, et al., 'Supplementary Data from ONC201 in Combination with Paxalisib for the Treatment of H3K27-Altered Diffuse Midline Glioma', American Association for Cancer Research (AACR) (2023)
|
|||||||
| Show 2 more others | ||||||||
Preprint (1 outputs)
| Year | Citation | Altmetrics | Link | |||||
|---|---|---|---|---|---|---|---|---|
| 2023 |
Duchatel R, Jackson E, Parackal S, Sun C, Daniel P, Mannan A, et al., 'PI3K/mTOR is a therapeutically targetable genetic dependency in diffuse intrinsic pontine glioma (2023)
|
|||||||
Grants and Funding
Summary
| Number of grants | 92 |
|---|---|
| Total funding | $19,176,750 |
Click on a grant title below to expand the full details for that specific grant.
20243 grants / $724,891
Taming free radicals to silence the epigenome of kinase active paediatric cancers. $591,891
Funding body: Cancer Australia
| Funding body | Cancer Australia |
|---|---|
| Project Team | Professor Matt Dun, Doctor Zacary Germon, Doctor Ryan Duchatel, Doctor Janis Chamberlain, Prof David Eisenstat, Associate Professor Jonathan Morris, Dr Iliya Dragutinovic |
| Scheme | Priority-driven Collaborative Cancer Research Scheme |
| Role | Lead |
| Funding Start | 2024 |
| Funding Finish | 2027 |
| GNo | G2300802 |
| Type Of Funding | C1500 - Aust Competitive - Commonwealth Other |
| Category | 1500 |
| UON | Y |
Utilising a translational nanopharmaceutics approach to improving drug delivery in diffuse intrinsic pontine glioma$113,000
Funding body: Mark Hughes Foundation
| Funding body | Mark Hughes Foundation |
|---|---|
| Project Team | Associate Professor Susan Hua, Miss Lauren Arms, Doctor Ryan Duchatel, Professor Matt Dun, Miss Evie Jackson |
| Scheme | Innovation Grant |
| Role | Investigator |
| Funding Start | 2024 |
| Funding Finish | 2025 |
| GNo | G2301481 |
| Type Of Funding | Scheme excluded from IGS |
| Category | EXCL |
| UON | Y |
HMRI Award for Mid-Career Research 2023$20,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2024 |
| Funding Finish | 2024 |
| GNo | G2400154 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
202316 grants / $6,000,886
The I-DIMENSIONS Project: Integrated DMG genomIc Methylomic EpigeNetic Spatial transcrIptomic prOteomic subtypiNg System$1,654,668
Funding body: Charlie Teo Foundation
| Funding body | Charlie Teo Foundation |
|---|---|
| Project Team | Professor Matt Dun, Professor Murray Cairns, Doctor Heather Lee, Doctor Sebastian Waszak, Dr Fa Valdes Mora, Professor Melissa Davies, Doctor Jordan Hansford, Prof Melissa Davies , Dr Jordan Hansford , Dr Sebastian Waszak |
| Scheme | More Data Grant |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2023 |
| GNo | G2300261 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
A chink in the armour of DMG tumours: Exploiting DMG-specific defence systems to improve response to treatment$1,200,000
Funding body: RUN DIPG
| Funding body | RUN DIPG |
|---|---|
| Project Team | Professor Matt Dun, Miss Evie Jackson, Doctor Ryan Duchatel, Miss Evangeline Jackson |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2026 |
| GNo | G2300913 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
ADDING DIMENSION TO DIFFUSE MIDLINE GLIOMA SUBTYPING – advanced tumour classification using a bioinformatics approach$1,000,000
Funding body: The Kinghorn Foundation
| Funding body | The Kinghorn Foundation |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | The Evie Poolman Research Gift |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2026 |
| GNo | G2300945 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Precision immunotherapeutic strategy for paediatric brain tumours$635,000
Funding body: Australian Lions Childhood Cancer Research Foundation
| Funding body | Australian Lions Childhood Cancer Research Foundation |
|---|---|
| Project Team | Professor Matt Dun, Doctor Ryan Duchatel, Doctor Pouya Faridi, Doctor Dilana Staudt Barreto |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2025 |
| GNo | G2300290 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Enhancing the recruitment of tumor infiltrating lymphocytes to improve responses to DMG therapies$525,432
Funding body: The Cure Starts Now Foundation
| Funding body | The Cure Starts Now Foundation |
|---|---|
| Project Team | Professor Matt Dun, Doctor Frank Alvaro, Dr Frank Alvaro, Dr Tiago Carvalheiro, Dr David Gallego Ortega, Dr Carl Koschmann, Prof Carl Koschmann, Fatima Mora, David Ortega, Dr Fa Valdes Mora, Dr Nicholas Vitanza, Dr Nicolas Vitanza, Prof Jasper van der Lugt |
| Scheme | DIPG/DMG Collaborative Research Grant |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2024 |
| GNo | G2200735 |
| Type Of Funding | C3500 – International Not-for profit |
| Category | 3500 |
| UON | Y |
DMG COMBATT 2.0: Diffuse Midline Glioma Combined Anti-Tumor Targeting 2.0$304,092
Funding body: The Cure Starts Now Foundation
| Funding body | The Cure Starts Now Foundation |
|---|---|
| Project Team | Doctor Ryan Duchatel, Professor Matt Dun, Professor Hubert Hondermarck, Professor Jonathan Morris, Professor Jonathan Morris |
| Scheme | Research Grant |
| Role | Investigator |
| Funding Start | 2023 |
| Funding Finish | 2024 |
| GNo | G2200705 |
| Type Of Funding | C3500 – International Not-for profit |
| Category | 3500 |
| UON | Y |
Doubling Down: Enhancing the therapeutic benefit of investigational drugs for diffuse midline glioma$150,000
Funding body: The Blackjack Foundation
| Funding body | The Blackjack Foundation |
|---|---|
| Project Team | Professor Matt Dun, Doctor Ryan Duchatel, Miss Evangeline Jackson |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2025 |
| GNo | G2300289 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
The RUN DIPG Warrior Jack Scholarship$135,312
Funding body: RUN DIPG
| Funding body | RUN DIPG |
|---|---|
| Project Team | Professor Matt Dun, Un-named Student |
| Scheme | Scholarship |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2026 |
| GNo | G2300606 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Targeting cholesterol biosynthesis for improved efficacy of hypomethylating agents in acute myeloid leukaemia$100,000
Funding body: Cure Cancer Australia Foundation
| Funding body | Cure Cancer Australia Foundation |
|---|---|
| Project Team | Doctor Danielle Bond, Professor Matt Dun, Doctor Anoop Enjeti, Doctor Heather Lee, Associate Professor Nikki Verrills |
| Scheme | Research Grant |
| Role | Investigator |
| Funding Start | 2023 |
| Funding Finish | 2023 |
| GNo | G2200647 |
| Type Of Funding | C1700 - Aust Competitive - Other |
| Category | 1700 |
| UON | Y |
Targeting insulin signalling to overcome resistance to venetoclax in acute myeloid leukaemia (AML)$100,000
Funding body: Cure Cancer Australia Foundation
| Funding body | Cure Cancer Australia Foundation |
|---|---|
| Project Team | Doctor Heather Murray, Dr Natasha Anstee, Doctor Natasha Anstee, Professor Matt Dun, Doctor Anoop Enjeti, Associate Professor Nikki Verrills, Prof Andrew Wei |
| Scheme | Research Grant |
| Role | Investigator |
| Funding Start | 2023 |
| Funding Finish | 2023 |
| GNo | G2200755 |
| Type Of Funding | C1700 - Aust Competitive - Other |
| Category | 1700 |
| UON | Y |
Evaluating promising anti-DMG therapies through enhanced collaboration$56,000
Funding body: RUN DIPG
| Funding body | RUN DIPG |
|---|---|
| Project Team | Doctor Ryan Duchatel, Doctor Dilana Staudt Barreto, Doctor Zacary Germon, Miss Evangeline Jackson, Ms Mika Persson, Mr Izac Findlay, Ms Mika Persson, Mr Izac Findlay, Mr Bryce Thomas, Professor Matt Dun, Mr Bryce Thomas |
| Scheme | Research Grant |
| Role | Investigator |
| Funding Start | 2023 |
| Funding Finish | 2024 |
| GNo | G2300438 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Establishment of a developmentally, genomically and immunologically relevant animal avatar of diffuse midline glioma$53,588
Funding body: Isabella and Marcus Foundation Limited
| Funding body | Isabella and Marcus Foundation Limited |
|---|---|
| Project Team | Doctor Ryan Duchatel, Professor Matt Dun |
| Scheme | Research Grant |
| Role | Investigator |
| Funding Start | 2023 |
| Funding Finish | 2023 |
| GNo | G2301063 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
Taming free radicals to increase response to standard-of-care treatments for children with high-risk leukaemias$30,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Doctor Zacary Germon, Doctor Heather Lee |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2023 |
| GNo | G2300163 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Establishment of a developmentally, genomically and immunologically relevant animal avatar of diffuse midline glioma$26,794
Funding body: Little Legs Foundation
| Funding body | Little Legs Foundation |
|---|---|
| Project Team | Doctor Ryan Duchatel, Professor Matt Dun |
| Scheme | Research Grant |
| Role | Investigator |
| Funding Start | 2023 |
| Funding Finish | 2024 |
| GNo | G2301067 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
Targeting the untargetable - targeting the mutant genes, proteins and pathways responsible for paediatric high-grade gliomas, to improve treatment recommendations for patients$25,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Mr Izac Findlay |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2023 |
| Funding Finish | 2023 |
| GNo | G2300692 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
External collaboration_Domestic_Barreto$5,000
Funding body: University of Newcastle
| Funding body | University of Newcastle |
|---|---|
| Project Team | Doctor Dilana Staudt Barreto, Professor Matt Dun |
| Scheme | External Collaboration Grant Scheme - Domestic |
| Role | Investigator |
| Funding Start | 2023 |
| Funding Finish | 2023 |
| GNo | G2300418 |
| Type Of Funding | Internal |
| Category | INTE |
| UON | Y |
20229 grants / $2,913,699
in vitro and in vivo assessment of Aptabio-19 against acute high-risk leukaemias including patient-derived xenograft modelling.$846,229
Funding body: AptaBio Therapeutics Inc
| Funding body | AptaBio Therapeutics Inc |
|---|---|
| Project Team | Professor Matt Dun, Doctor Abdul Mannan, Associate Professor Geoffry De Iuliis |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2023 |
| GNo | G2200170 |
| Type Of Funding | C3400 – International For Profit |
| Category | 3400 |
| UON | Y |
The Wish Lab$814,123
Funding body: RUN DIPG
| Funding body | RUN DIPG |
|---|---|
| Project Team | Professor Matt Dun, Doctor Ryan Duchatel |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2022 |
| GNo | G2200206 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
INTREPID – trINitarian TReatment tEmplate to imProve the survIval of DIPG patients$477,533
Funding body: Little Legs Foundation
| Funding body | Little Legs Foundation |
|---|---|
| Project Team | Professor Matt Dun, Mr Bryce Thomas |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2026 |
| GNo | G2201086 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
Pharmaco-phospho-proteo-genomics of paediatric high-grade glioma$300,000
Funding body: The Kids' Cancer Project
| Funding body | The Kids' Cancer Project |
|---|---|
| Project Team | Professor Matt Dun, Dr William Lo, Doctor Frank Alvaro, Dr Mark Dexter, Doctor Mitchell Hansen, Dr William Lo |
| Scheme | HGG brain cancers |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2024 |
| GNo | G2101095 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
Pharmaco-phospho-proteo-genomics of paediatric high-grade glioma$258,000
Funding body: RUN DIPG
| Funding body | RUN DIPG |
|---|---|
| Project Team | Professor Matt Dun, Dr Frank Alvaro, Mr Izac Findlay, Associate Professor Mitchell Hansen, Dr William Lo |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2025 |
| GNo | G2200066 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Progressing promising anti cancer strategies for the treatment of high-risk paediatric cancer$100,000
Funding body: Maitland Cancer Appeal Committee Incorporated
| Funding body | Maitland Cancer Appeal Committee Incorporated |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Funding |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2022 |
| GNo | G2200512 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Understanding immune system involvement in diffuse midline glioma$54,538
Funding body: Liv like a Unicorn Foundation
| Funding body | Liv like a Unicorn Foundation |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2022 |
| GNo | G2200389 |
| Type Of Funding | C3500 – International Not-for profit |
| Category | 3500 |
| UON | Y |
Enhanced evaluation of disease drivers in high-risk paediatric cancers$50,000
Funding body: The Blackjack Foundation
| Funding body | The Blackjack Foundation |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2022 |
| GNo | G2200718 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Developing analysis techniques to understand disease drivers and therapeutic vulnerability in diffuse intrinsic pontine glioma$13,276
Funding body: Schwab Charitable
| Funding body | Schwab Charitable |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | The Hirsch Family Funderpants |
| Role | Lead |
| Funding Start | 2022 |
| Funding Finish | 2022 |
| GNo | G2200293 |
| Type Of Funding | C3500 – International Not-for profit |
| Category | 3500 |
| UON | Y |
202111 grants / $1,488,457
Phosphoproteogenomic characterisation of diffuse intrinsic pontine glioma (DIPG)$520,493
Funding body: RUN DIPG
| Funding body | RUN DIPG |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2021 |
| GNo | G2100084 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
COMBATT DMG: Combined anti-tumour targeting of diffuse midline glioma$203,522
Funding body: The Cure Starts Now Foundation
| Funding body | The Cure Starts Now Foundation |
|---|---|
| Project Team | Professor Matt Dun, Dr Jason Cain, Doctor Ryan Duchatel, Jason Cain |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2022 |
| GNo | G2000780 |
| Type Of Funding | C3500 – International Not-for profit |
| Category | 3500 |
| UON | Y |
In vivo evaluation of paxalisib for the treatment of diffuse midline glioma$198,000
Funding body: Kazia Therapeutics Limited
| Funding body | Kazia Therapeutics Limited |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2022 |
| GNo | G2101225 |
| Type Of Funding | C3100 – Aust For Profit |
| Category | 3100 |
| UON | Y |
Rapid advancement of central nervous system active therapies to clinical trial$125,978
Funding body: Pacific Pediatric Neuro-Oncology Consortium Foundation
| Funding body | Pacific Pediatric Neuro-Oncology Consortium Foundation |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2021 |
| GNo | G2100894 |
| Type Of Funding | C3600 - International Philanthropy |
| Category | 3600 |
| UON | Y |
Cracking the Code: The launch of a genomic, epigenetic and proteomic pre-clinical platform to improve the treatment of paediatric leukemias$122,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills, Doctor Heather Lee, Doctor Janis Chamberlain, Doctor Frank Alvaro, Doctor Anoop Enjeti, Associate Professor Kathryn Skelding, Doctor Lisa Lincz, Doctor Abdul Mannan, Doctor Heather Murray, Kristy McCarthy, Elizabeth Heskett, Paola Baeza, Kathleen Irish |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2021 |
| GNo | G2001337 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Neoantigen immunopeptidomics for the development of immunotherapies for the treatment of diffuse intrinsic pontine glioma (DIPG) $108,251
Funding body: RUN DIPG
| Funding body | RUN DIPG |
|---|---|
| Project Team | Professor Matt Dun, Professor Jay Horvat, Associate Professor Martin Larsen, Associate Professor Luke Hesson, Associate Professor Mark Cowley |
| Scheme | Scholarship |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2024 |
| GNo | G2001148 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Optimising novel central nervous system active drugs for the treatment of paediatric high-grade gliomas$100,000
Funding body: Strategic Group
| Funding body | Strategic Group |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2024 |
| GNo | G2100737 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Unravelling genomic heterogeneity of diffuse intrinsic pontine glioma$52,250
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Doctor Ryan Duchatel |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2023 |
| GNo | G2100434 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Targeting NAPDH oxidase enzymes to improve the treatment of high-risk paediatric leukaemias $30,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Doctor Frank Alvaro, Doctor Abdul Mannan, Doctor Janis Chamberlain |
| Scheme | HCRF Project Grant |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2021 |
| GNo | G2100141 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Edie’s Kindness Research Donation$18,236
Funding body: Craig and Lois Jackson - Edie's Kindness Project
| Funding body | Craig and Lois Jackson - Edie's Kindness Project |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Donation |
| Role | Lead |
| Funding Start | 2021 |
| Funding Finish | 2021 |
| GNo | G2101278 |
| Type Of Funding | C3600 - International Philanthropy |
| Category | 3600 |
| UON | Y |
The utility of specialised Blood Collection Tubes (STRECK-BCT) for the detection of CEA in colorectal cancer patients$9,727
Funding body: John Hunter Hospital Charitable Trust
| Funding body | John Hunter Hospital Charitable Trust |
|---|---|
| Project Team | Doctor Abdul Mannan, Professor Matt Dun, Dr Stanley Chen |
| Scheme | Research Grant |
| Role | Investigator |
| Funding Start | 2021 |
| Funding Finish | 2021 |
| GNo | G2001170 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
20204 grants / $2,775,089
Utilising male fertility as a biomarker of health to understand the biological effects of PFAS$1,346,912
Funding body: NHMRC (National Health & Medical Research Council)
| Funding body | NHMRC (National Health & Medical Research Council) |
|---|---|
| Project Team | Professor Brett Nixon, Associate Professor Brett Turner, Associate Professor Brett Turner, Associate Professor Geoffry De Iuliis, Associate Professor Geoffry De Iuliis, Dr Bradley Clarke, Doctor Shaun Roman, Doctor Shaun Roman, Professor Matt Dun, Professor Matt Dun, Doctor Andy Eamens, Doctor Andy Eamens, A/Prof Mark Green, A/Prof Mark Green, Dr Bradley Clarke, Miss Leah Calvert |
| Scheme | Targeted Call for Research - Per and Poly-Fluoroalkylated Substances (PFAS) |
| Role | Investigator |
| Funding Start | 2020 |
| Funding Finish | 2023 |
| GNo | G1900626 |
| Type Of Funding | C1100 - Aust Competitive - NHMRC |
| Category | 1100 |
| UON | Y |
Mechanistic and translational studies to improve the outcomes of high-risk paediatric cancers$644,845
Funding body: NHMRC (National Health & Medical Research Council)
| Funding body | NHMRC (National Health & Medical Research Council) |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Investigator Grant |
| Role | Lead |
| Funding Start | 2020 |
| Funding Finish | 2024 |
| GNo | G1801489 |
| Type Of Funding | C1100 - Aust Competitive - NHMRC |
| Category | 1100 |
| UON | Y |
Unlocking oncogene addition to identify synergistic treatment targets for the treatment of diffuse intrinsic pontine glioma$456,943
Funding body: Michael Mosier Defeat DIPG Foundation
| Funding body | Michael Mosier Defeat DIPG Foundation |
|---|---|
| Project Team | Professor Matt Dun, Doctor Ryan Duchatel |
| Scheme | Defeat DIPG ChadTough New Investigator Grant |
| Role | Lead |
| Funding Start | 2020 |
| Funding Finish | 2022 |
| GNo | G1901112 |
| Type Of Funding | C3500 – International Not-for profit |
| Category | 3500 |
| UON | Y |
Harnessing the power from within: Neoantigen immunopeptidomics for the development of immunotherapies for the treatment of diffuse intrinsic pontine glioma (DIPG)$326,389
Funding body: Little Legs Foundation
| Funding body | Little Legs Foundation |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Mark Cowley, Associate Professor Luke Hesson |
| Scheme | More Data Thesis |
| Role | Lead |
| Funding Start | 2020 |
| Funding Finish | 2022 |
| GNo | G2000193 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
20199 grants / $484,656
New horizons. Therapeutic applications for medicinal cannabis in the treatment of brain cancer.$190,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Dr Kelly McKelvey, Doctor Adjanie Patabendige, Doctor Ameha Woldu, Doctor Mengna Chi, Doctor Craig Gedye |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2019 |
| Funding Finish | 2020 |
| GNo | G1900131 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Blueprint to a cure - Mapping the proteogenomic architecture of diffuse intrinsic pontine gliomas$103,535
Funding body: Tour De Cure
| Funding body | Tour De Cure |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Pioneering Research Grant |
| Role | Lead |
| Funding Start | 2019 |
| Funding Finish | 2020 |
| GNo | G1901190 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
An integrated proteomics approach to identify novel targets for treatment of Acute Myeloid Leukaemia$58,181
Funding body: Takeda Pharmaceutical Company Limited
| Funding body | Takeda Pharmaceutical Company Limited |
|---|---|
| Project Team | Associate Professor Nikki Verrills, Professor Matt Dun, Professor Richard D’Andrea, Dr David Ross |
| Scheme | COCKPI-T Australia |
| Role | Investigator |
| Funding Start | 2019 |
| Funding Finish | 2019 |
| GNo | G1801222 |
| Type Of Funding | C3400 – International For Profit |
| Category | 3400 |
| UON | Y |
Characterisation of medicinal cannabis$52,440
Funding body: Department of Industry, Innovation and Science
| Funding body | Department of Industry, Innovation and Science |
|---|---|
| Project Team | Professor Matt Dun, Doctor Mengna Chi, Doctor Ameha Woldu, Doctor Ameha Woldu |
| Scheme | Entrepreneurs' Programme: Innovation Connections |
| Role | Lead |
| Funding Start | 2019 |
| Funding Finish | 2019 |
| GNo | G1801362 |
| Type Of Funding | C3100 – Aust For Profit |
| Category | 3100 |
| UON | Y |
Targeting BCL6 for the treatment of relapsed FLT3 mutant acute myeloid leukaemia (AML)$30,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Miss Dilana Staudt Barreto, Doctor Frank Alvaro, Doctor Kristy McCarthy, Dr Charles de Bock |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2019 |
| Funding Finish | 2020 |
| GNo | G1901490 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Moving safe and well-tolerated therapies from the bench to the clinic for the treatment of childhood brain cancer$26,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Doctor Ryan Duchatel, Doctor Frank Alvaro, Dr Javad Nazarian, Dr Michelle Monje |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2019 |
| Funding Finish | 2020 |
| GNo | G1901488 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Preclinical research into the potential applications of GDC-0084 in diffuse intrinsic pontine glioma (DIPG)$10,000
Funding body: Kazia Therapeutics Limited
| Funding body | Kazia Therapeutics Limited |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor David Ziegler, Doctor Heather Murray, Doctor Ryan Duchatel, Doctor Frank Alvaro |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2019 |
| Funding Finish | 2019 |
| GNo | G1801161 |
| Type Of Funding | C3100 – Aust For Profit |
| Category | 3100 |
| UON | Y |
2019 NSW Premier's Awards for Outstanding Cancer Research — Outstanding Cancer Research Fellow: Dr Matthew Dun$10,000
Funding body: Cancer Institute NSW
| Funding body | Cancer Institute NSW |
|---|---|
| Project Team | Professor Matt Dun, Professor Matt Dun |
| Scheme | Premier's Award for Outstanding Cancer Researcher of the Year |
| Role | Lead |
| Funding Start | 2019 |
| Funding Finish | 2019 |
| GNo | G1901438 |
| Type Of Funding | C2400 – Aust StateTerritoryLocal – Other |
| Category | 2400 |
| UON | Y |
Kyratec PCR machine$4,500
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Associate Professor Nikki Verrills, Doctor Severine Roselli Dayas, Professor Matt Dun |
| Scheme | Early and Mid-Career Equipment Grant |
| Role | Investigator |
| Funding Start | 2019 |
| Funding Finish | 2019 |
| GNo | G1900111 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
201815 grants / $1,282,414
Elucidating the role of epididymosomes in the transfer of fertility-modulating proteins and regulatory classes of RNA to maturing spermatozoa$547,155
Funding body: NHMRC (National Health & Medical Research Council)
| Funding body | NHMRC (National Health & Medical Research Council) |
|---|---|
| Project Team | Professor Brett Nixon, Professor Matt Dun |
| Scheme | Project Grant |
| Role | Investigator |
| Funding Start | 2018 |
| Funding Finish | 2020 |
| GNo | G1700434 |
| Type Of Funding | C1100 - Aust Competitive - NHMRC |
| Category | 1100 |
| UON | Y |
Novel therapeutic targets and methods for detection of high grade brain cancers$197,728
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2021 |
| GNo | G1800874 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Proteomic architecture of diffuse pontine intrinsic glioma$100,000
Funding body: McDonald Jones Charitable Foundation
| Funding body | McDonald Jones Charitable Foundation |
|---|---|
| Project Team | Professor Matt Dun, Doctor Frank Alvaro, Doctor Ryan Duchatel, Doctor Heather Murray, Associate Professor David Ziegler |
| Scheme | Postdoctoral fellowship |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2020 |
| GNo | G1801130 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Preclinical evaluation of novel therapies for the treatment of diffuse intrinsic pontine glioma$100,000
To support the development of new treatments for DIPG Dr Dun's laboratory in collaboration with A/Prof Zieglers laboratory will perform preclinical testing of two new drugs.
Funding body: McDonald Jones Charitable Foundation
| Funding body | McDonald Jones Charitable Foundation |
|---|---|
| Project Team | Dr Matt Dun, A/Prof David Ziegler (UNSW) |
| Scheme | Preclinical testing |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2020 |
| GNo | |
| Type Of Funding | C3120 - Aust Philanthropy |
| Category | 3120 |
| UON | N |
Investigation of disease markers in asymptomatic patients with colorectal carcinoma or colonic adenomas$88,629
Funding body: Streck
| Funding body | Streck |
|---|---|
| Project Team | Professor Matt Dun, Doctor Peter Pockney, Professor Rodney Scott |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2022 |
| GNo | G1800707 |
| Type Of Funding | C3400 – International For Profit |
| Category | 3400 |
| UON | Y |
Capillary Flow Two Dimensional High Pressure Liquid Chromatography (HPLC) system$75,761
Funding body: NHMRC (National Health & Medical Research Council)
| Funding body | NHMRC (National Health & Medical Research Council) |
|---|---|
| Project Team | Professor Brett Nixon, Professor Matt Dun, Associate Professor Nikki Verrills, Professor Hubert Hondermarck, Associate Professor Mark Baker, Doctor Elizabeth Bromfield |
| Scheme | Equipment Grant |
| Role | Investigator |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1800470 |
| Type Of Funding | Scheme excluded from IGS |
| Category | EXCL |
| UON | Y |
Non-invasive detection of DIPG specific DNA and protein using sequential blood collections$57,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Doctor Muhammad Fairuz Jamaluddin, Doctor Ryan Duchatel, Doctor Frank Alvaro |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2020 |
| GNo | G1801235 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Enhancing the efficacy of new inhibitors targeting the PI3K–AKT–mTOR signalling axis for the treatment of high-grade diffuse intrinsic pontine gliomas (DIPG)$30,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Doctor Ryan Duchatel, Doctor Adjanie Patabendige |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1801386 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Targeting Reactive Oxygen Species Generation as a Novel Treatment Target in Acute Myeloid Leukaemia - RA support$25,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Doctor Jonathan Sillar, Doctor Anoop Enjeti, Associate Professor Nikki Verrills |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1800399 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
Targeting DNA repair for the improved treatment of blood cancers$20,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1800611 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Proteomic Profiling of Chemoresistant Acute Lymphoblastic Leukemia-Derived Exosomes$10,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Dr Kara Perrow, Associate Professor Nikki Verrills, Professor Maria Kavallaris, Doctor Muhammad Fairuz Jamaluddin |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1801240 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Preclinical testing of new treatments for DIPG$10,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Doctor Frank Alvaro, Professor Chris Dayas, Doctor Ryan Duchatel, Associate Professor David Ziegler |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1801241 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Building international collaborations for DIPG research$10,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Doctor Ryan Duchatel, Professor Matt Dun |
| Scheme | Jennie Thomas Medical Research Travel Grant |
| Role | Investigator |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1801371 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Targeting oncogenic signalling in DIPG using drugs that cross the blood brain barrier.$7,000
Funding body: Australian Communities Foundation
| Funding body | Australian Communities Foundation |
|---|---|
| Project Team | Professor Matt Dun, Doctor Ryan Duchatel |
| Scheme | Isabella and Marcus Paediatric Brainstem Tumour Fund |
| Role | Lead |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1800977 |
| Type Of Funding | C3200 – Aust Not-for Profit |
| Category | 3200 |
| UON | Y |
Jennie Thomas Medical Research Travel Grant - Nikita Panicker$4,141
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Miss Nikita Panicker, Associate Professor Nikki Verrills, Doctor Severine Roselli Dayas, Professor Matt Dun |
| Scheme | Jennie Thomas Medical Research Travel Grant |
| Role | Investigator |
| Funding Start | 2018 |
| Funding Finish | 2018 |
| GNo | G1800024 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
20179 grants / $953,900
Anticancer properties of cannabis oil and its individual components$400,000
Funding body: Australian Natural Therapeutics Group Pty Ltd
| Funding body | Australian Natural Therapeutics Group Pty Ltd |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2017 |
| Funding Finish | 2018 |
| GNo | G1700581 |
| Type Of Funding | C3100 – Aust For Profit |
| Category | 3100 |
| UON | Y |
Anticancer Properties of Low THC Hemp and its components$210,419
Funding body: Cancer Institute NSW
| Funding body | Cancer Institute NSW |
|---|---|
| Project Team | Doctor Mengna Chi, Professor Matt Dun, Associate Professor Jenny Schneider |
| Scheme | Early Career Fellowship |
| Role | Investigator |
| Funding Start | 2017 |
| Funding Finish | 2024 |
| GNo | G1700667 |
| Type Of Funding | C2300 – Aust StateTerritoryLocal – Own Purpose |
| Category | 2300 |
| UON | Y |
Targeting activated signalling and oxidative stress pathways in acute myeloid leukaemia$150,000
Funding body: Cancer Institute NSW
| Funding body | Cancer Institute NSW |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Early Career Fellowship |
| Role | Lead |
| Funding Start | 2017 |
| Funding Finish | 2020 |
| GNo | G1600806 |
| Type Of Funding | C2300 – Aust StateTerritoryLocal – Own Purpose |
| Category | 2300 |
| UON | Y |
Developing new treatments for resistance in acute myeloid leukaemia$57,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2017 |
| Funding Finish | 2020 |
| GNo | G1701507 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
HMRI MRSP Special Infrastructure Scheme Chemidoc$56,600
Funding body: HMRI
| Funding body | HMRI |
|---|---|
| Project Team | Matt Dun, Hubert Hondermarck, Murray Cairns, Brett Nixon, Phillip Dickson, Nikki Verrills |
| Scheme | HMRI |
| Role | Lead |
| Funding Start | 2017 |
| Funding Finish | 2018 |
| GNo | |
| Type Of Funding | Aust Competitive - Non Commonwealth |
| Category | 1NS |
| UON | N |
Receptor tyrosine kinase mutations in acute myeloid leukaemia promote PP2A and p53 inhibition through the phosphorylation of SBDS$27,500
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2017 |
| Funding Finish | 2017 |
| GNo | G1700272 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Determining the mechanisms underpinning leukaemic transformation for children suffering from Shwachman-Diamond Syndrome (SDS)$25,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills, Doctor Bryony Ross, Doctor Anoop Enjeti, Dr Jeremy Robertson |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2017 |
| Funding Finish | 2017 |
| GNo | G1701574 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Targeting a tumour suppressor for new cancer therapies $24,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Associate Professor Nikki Verrills, Professor Matt Dun, Doctor Severine Roselli Dayas |
| Scheme | Project Grant |
| Role | Investigator |
| Funding Start | 2017 |
| Funding Finish | 2018 |
| GNo | G1700588 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Jennie Thomas Medical Research Travel Grant$3,381
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Doctor David Skerrett-Byrne, Professor Phil Hansbro, Professor Matt Dun |
| Scheme | Jennie Thomas Medical Research Travel Grant |
| Role | Investigator |
| Funding Start | 2017 |
| Funding Finish | 2017 |
| GNo | G1701070 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
20161 grants / $100,000
Targeting DNA repair for the improved treatment of blood cancers$100,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2016 |
| Funding Finish | 2016 |
| GNo | G1601127 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
20155 grants / $1,031,250
High resolution fourier transform mass spectrometry platform for the discovery of novel cancer biomarkers and drug targets using label-free and isobaric-tagged approaches for quantitative proteomics$500,000
Funding body: Cancer Instititue NSW
| Funding body | Cancer Instititue NSW |
|---|---|
| Project Team | X. Dong Zhang, M.D. Dun, J. Martin, H. Hondermarck, R.J. Aitken, N.M. Verrills, P. Tanwar, R. Scott, M. Kavallaris (UNSW), D. Saunders (USyd) |
| Scheme | Research Equipment Grant |
| Role | Lead |
| Funding Start | 2015 |
| Funding Finish | 2016 |
| GNo | |
| Type Of Funding | Aust Competitive - Non Commonwealth |
| Category | 1NS |
| UON | N |
Advanced Technical Support for Oncology Single Cell Analysis Technologies$300,000
Funding body: Cancer Institute NSW
| Funding body | Cancer Institute NSW |
|---|---|
| Project Team | Professor Rodney Scott, Professor Xu Dong Zhang, Professor Hubert Hondermarck, Conjoint Professor Stephen Ackland, Doctor Craig Gedye, Professor Pradeep Tanwar, Doctor Chen Chen Jiang, Professor Matt Dun, Professor Paul de Souza, Associate Professor Kevin Spring, Dr Tao Liu |
| Scheme | Research Infrastructure Grants |
| Role | Investigator |
| Funding Start | 2015 |
| Funding Finish | 2018 |
| GNo | G1500824 |
| Type Of Funding | C2300 – Aust StateTerritoryLocal – Own Purpose |
| Category | 2300 |
| UON | Y |
High resolution fourier transform mass spectrometry platform for the discovery of novel cancer biomarkers and drug targets using label-free and isobaric-tagged approaches for quantitative proteomics.$196,250
Funding body: University of Newcastle
| Funding body | University of Newcastle |
|---|---|
| Project Team | Professor Xu Dong Zhang, Professor Matt Dun, Professor Jennifer Martin, Professor Hubert Hondermarck, Distinguished Emeritus Professor John Aitken, Associate Professor Nikki Verrills, Professor Pradeep Tanwar, Professor Rodney Scott, Professor Maria Kavallaris, Dr Darren Saunders |
| Scheme | Equipment Grant |
| Role | Investigator |
| Funding Start | 2015 |
| Funding Finish | 2015 |
| GNo | G1500935 |
| Type Of Funding | Internal |
| Category | INTE |
| UON | Y |
Identification of better diagnostic tools and new treatment options for children suffering from Shwachman-Diamond Syndrome (SDS)$25,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills |
| Scheme | Research Grant |
| Role | Lead |
| Funding Start | 2015 |
| Funding Finish | 2015 |
| GNo | G1500757 |
| Type Of Funding | Grant - Aust Non Government |
| Category | 3AFG |
| UON | Y |
Jennie Thomas Medical Research Travel Grant$10,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Jennie Thomas Medical Research Travel Grant |
| Role | Lead |
| Funding Start | 2015 |
| Funding Finish | 2015 |
| GNo | G1501429 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
20146 grants / $1,289,412
Identifying novel therapeutic targets for the treatment of Acute Myeloid Leukaemia$600,000
Funding body: Cancer Institute NSW
| Funding body | Cancer Institute NSW |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills, Associate Professor Martin Larsen |
| Scheme | Early Career Fellowship |
| Role | Lead |
| Funding Start | 2014 |
| Funding Finish | 2016 |
| GNo | G1300952 |
| Type Of Funding | Other Public Sector - State |
| Category | 2OPS |
| UON | Y |
Tetraspanin CD9; more than just an exosome marker - A novel biomarker to target for prostate cancer$400,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Doctor Jude Weidenhofer, Associate Professor Kathryn Skelding, Professor Matt Dun, Ms Belinda Goldie, Doctor Danielle Bond |
| Scheme | Project Grant |
| Role | Investigator |
| Funding Start | 2014 |
| Funding Finish | 2017 |
| GNo | G1400921 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Visualisation of microparticles for development of biomarkers and targeted drug delivery mechanisms$125,199
Funding body: Cancer Institute NSW
| Funding body | Cancer Institute NSW |
|---|---|
| Project Team | Professor Christopher Scarlett, Associate Professor Kathryn Skelding, Doctor Jude Weidenhofer, Professor Matt Dun, Associate Professor Kelly Kiejda, Professor Adam McCluskey, Doctor Elham Sadeqzadeh, Professor Hubert Hondermarck, Doctor Rick Thorne, Professor Rodney Scott |
| Scheme | Research Equipment Grant |
| Role | Investigator |
| Funding Start | 2014 |
| Funding Finish | 2014 |
| GNo | G1400627 |
| Type Of Funding | Other Public Sector - State |
| Category | 2OPS |
| UON | Y |
JuLI Stage $71,674
Funding body: NHMRC (National Health & Medical Research Council)
| Funding body | NHMRC (National Health & Medical Research Council) |
|---|---|
| Project Team | Professor Pradeep Tanwar, Professor Eileen McLaughlin, Emeritus Professor Robin Callister, Professor Xu Dong Zhang, Professor Murray Cairns, Professor Brett Nixon, Professor Hubert Hondermarck, Associate Professor Phillip Dickson, Associate Professor Nikki Verrills, Professor Matt Dun, Doctor Jessie Sutherland, Doctor Janani Kumar, Professor Jay Horvat, Associate Professor Susan Hua, Prof LIZ Milward, Associate Professor Estelle Sontag, Professor Dirk Van Helden, Doctor Janet Bristow, Doctor Jean-Marie Sontag |
| Scheme | Equipment Grant |
| Role | Investigator |
| Funding Start | 2014 |
| Funding Finish | 2014 |
| GNo | G1500860 |
| Type Of Funding | Other Public Sector - Commonwealth |
| Category | 2OPC |
| UON | Y |
Identifying novel therapeutic targets for the treatment of Acute Myeloid Leukaemia$67,539
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2014 |
| Funding Finish | 2017 |
| GNo | G1301353 |
| Type Of Funding | C3300 – Aust Philanthropy |
| Category | 3300 |
| UON | Y |
Defining the role of shwachman-bodien diamond syndrome protein (SBDS) in PP2A inhibition in acute myeloid leukaemia (AML).$25,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2014 |
| Funding Finish | 2014 |
| GNo | G1301442 |
| Type Of Funding | Grant - Aust Non Government |
| Category | 3AFG |
| UON | Y |
20133 grants / $32,096
Ultra-Low Temperature Cryogenic Freezer$24,596
Funding body: NHMRC (National Health & Medical Research Council)
| Funding body | NHMRC (National Health & Medical Research Council) |
|---|---|
| Project Team | Doctor Jude Weidenhofer, Doctor Rick Thorne, Associate Professor Kathryn Skelding, Associate Professor Nikki Verrills, Professor Pradeep Tanwar, Associate Professor Phillip Dickson, Professor Murray Cairns, Professor Hubert Hondermarck, Professor Xu Dong Zhang, Associate Professor Estelle Sontag, Doctor Chen Chen Jiang, Prof LIZ Milward, Doctor Jean-Marie Sontag, Associate Professor Paul Tooney, Doctor Severine Roselli Dayas, Professor Matt Dun, Professor Chris Dayas, Doctor Lin Kooi Ong, Professor Dirk Van Helden, Mr Ben Copeland, Doctor Gabrielle Briggs, Emeritus Professor Leonie Ashman, Emeritus Professor John Rostas |
| Scheme | Equipment Grant |
| Role | Investigator |
| Funding Start | 2013 |
| Funding Finish | 2013 |
| GNo | G1201189 |
| Type Of Funding | Other Public Sector - Commonwealth |
| Category | 2OPC |
| UON | Y |
Proteomics of Cancer$6,000
Funding body: Hunter Medical Research Institute
| Funding body | Hunter Medical Research Institute |
|---|---|
| Project Team | Professor Matt Dun, Associate Professor Nikki Verrills, Associate Professor Anoop Enjeti |
| Scheme | Project Grant |
| Role | Lead |
| Funding Start | 2013 |
| Funding Finish | 2013 |
| GNo | G1300054 |
| Type Of Funding | Contract - Aust Non Government |
| Category | 3AFC |
| UON | Y |
American Association of Cancer Research, Washington Convention Centre Washington DC USA, 6 - 10 April 2012$1,500
Funding body: University of Newcastle - Faculty of Health and Medicine
| Funding body | University of Newcastle - Faculty of Health and Medicine |
|---|---|
| Project Team | Professor Matt Dun |
| Scheme | Travel Grant |
| Role | Lead |
| Funding Start | 2013 |
| Funding Finish | 2014 |
| GNo | G1201225 |
| Type Of Funding | Internal |
| Category | INTE |
| UON | Y |
20121 grants / $100,000
Hunter Translational Cancer Research Fellowship$100,000
Funding body: Cancer Instititue NSW
| Funding body | Cancer Instititue NSW |
|---|---|
| Scheme | ... |
| Role | Lead |
| Funding Start | 2012 |
| Funding Finish | 2012 |
| GNo | |
| Type Of Funding | Aust Competitive - Non Commonwealth |
| Category | 1NS |
| UON | N |
Research Supervision
Number of supervisions
Current Supervision
| Commenced | Level of Study | Research Title | Program | Supervisor Type |
|---|---|---|---|---|
| 2023 | PhD | Improving Drug Delivery Into The Brain | PhD (Pharmacy), College of Health, Medicine and Wellbeing, The University of Newcastle | Co-Supervisor |
| 2023 | PhD | Development of novel CAR T Therapies for Children Diagnosed with Central Nervous System Cancers | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
| 2022 | PhD | Pharmaco-Phospho-Proteo-Genomics of Paediatric High-Grade Glioma | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
| 2022 | PhD | Neoantigen Immunopeptidomics for the Development of Immunotherapies for the Treatment of Diffuse Intrinsic Pontine Glioma | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
| 2020 | PhD | Investigating the Effect of Hypoxia on Hypomethylating Agent Efficacy and Downstream Transcriptional Implications in Acute Myeloid Leukaemia | PhD (Medical Genetics), College of Health, Medicine and Wellbeing, The University of Newcastle | Co-Supervisor |
| 2018 | PhD | Investigation into the Anti-Acute Myeloid Leukaemia and Complementary Health Benefits of Medicinal Cannabis | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
| 2017 | PhD | Molecular Characterisation of Treatment Resistance in Acute Myeloid Leukaemia. | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
| 2016 | PhD | Reactive Oxygen Species Promoted Leukaemogenesis; Altered Signalling Pathways as New Drug Targets for the Improved Treatment of Acute Myeloid Leukaemia | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
Past Supervision
| Year | Level of Study | Research Title | Program | Supervisor Type |
|---|---|---|---|---|
| 2024 | PhD | Multiomic Characterisation of Cellular Signalling Regulated by PP2A-B55a in Breast Cancer and Development | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Co-Supervisor |
| 2023 | PhD | Molecular Characterisation of Oncogenic Signalling Networks to Develop Treatment Strategies for Diffuse Intrinsic Pontine Glioma | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
| 2022 | PhD | Targeting Reactive Oxygen Species for the Treatment of Acute Myeloid Leukaemia | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
| 2022 | PhD | Functional Role of Protein Phosphatase 2A and its Regulatory Subunit B55a in Development and Breast Cancer | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Co-Supervisor |
| 2022 | PhD | Molecular and Phosphoproteomic Characterisation of FLT3 Inhibitor Resistance in Acute Myeloid Leukaemia | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Principal Supervisor |
| 2021 | PhD | Proteomic analysis of neuroproteins in pancreatic cancer | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Co-Supervisor |
| 2020 | PhD | Identifying Novel Therapeutic Targets for the Treatment of Acute Myeloid Leukaemia | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Co-Supervisor |
| 2019 | PhD | Deep Time-Resolved Proteomic and Phosphoproteomic Profiling of Cigarette Smoke-Induced Chronic Obstructive Pulmonary Disease | PhD (Immunology & Microbiol), College of Health, Medicine and Wellbeing, The University of Newcastle | Co-Supervisor |
| 2019 | PhD | The Functional Role of PPP2R2A in Luminal Breast Cancer | PhD (Medical Biochemistry), College of Health, Medicine and Wellbeing, The University of Newcastle | Co-Supervisor |
| 2017 | Honours |
Characterisation of low Δ9-tetrahydrocannabinol containing medicinal cannabis for the treatment of acute myeloid leukaemia. <span style="font-size:12.0pt;font-family:'Arial',sans-serif;">Acute Myeloid Leukaemia (AML) carries a 5-year survival rate of 24%. Current treatments include high dose chemotherapy and bone marrow transplantation; however development of treatment resistance and relapse is common. Cannabis has been used for thousands of years as a traditional medicine and was introduced into western medicine in the nineteenth century. More recently, research into medicinal cannabis has focused on traditional high-THC containing products. Low-THC cannabis is legally defined as having less than 1% Δ9-THC and therefore, does not have the phsychotropic effects associated with tradtional cannabis. The aims of this research was to determine whether low-THC cannabis extracts were effective at reducing the survival of AML cells that are sensitive to new precision medicines and AML cells that have formed resistance to these therapies whilst determining whether low-THC cannabis effects the survival of normal healthy cells of the immune system. Two AML cell lines harbouring recurring genetic mutations were used and both show sensitivity to our crude low-THC cannabis extracts. To determine which cannabinoid or unknown chemicals was responsible for the cytotoxic effects we showed, High Performance Liquid Chromatography with Ultraviolet visible light detection (HPLC-UV Vis) was used. During this investigation we have optimised a HPLC-UV Vis technique that resolves thirteen common cannabinoids found in cannabis extracts and used this to analyse what was present in our low-THC containing cannabis extracts. Using various extraction techniques coupled to cytotoxic analysis and HPLC-UV, we have discovered that cannabidiol (CBD) and cannabichromene (CBC) are potentially invovled at inducing cell death of AML cells. Additionally, we have identified an unknown compound whose concentration in each of the extracts reflects the cytotoxicity seen. Future research is still necessary to analyse the unknown compounds and individual activity of compounds, but the research described herein reveals that low-THC containing cannabis is effective at killing AML cells.</span> |
Pharmacy, Faculty of Health and Medicine, University of Newcastle | Principal Supervisor |
| 2017 | Honours |
Targeting Oxidative Stress for the Improved Treatment of Acute Myeloid Leukaemia <p style="text-align:justify;line-height:150%;"><span style="font-family:Arial,Helvetica,sans-serif;font-size:small;">Treatment of acute myeloid leukaemia (AML) has remained unchanged over the last 35 years, with standard-of-care chemotherapeutics causing significant side effects and treatment induced death. AML patients often develop resistance to therapies, causing the formation of an even more aggressive untreatable disease that rapidly leads to death. Interest in reactive oxygen species has grown as studies have revealed that high levels of Reactive oxygen species (ROS) are seen in almost all cancer types, including AML. Despite this, the physiological and pathophysiological role that ROS plays in AML has widely remained unknown. It is a possibility that the altered redox environment initiated by the processes of cellular transformation may in fact drive leukaemogenesis. We believe this is through oxidative modifications to crucial regulators of cellular signalling, as well as acquired insults to nucleic acids and lipids, driving increased growth, survival and genomic instability. The altered redox environment may in fact be the cause for the development of drug resistance that is commonly seen during the treatment of an AML patient. These data raise the intriguing possibility of whether ROS may be a novel target for the treatment of AML.</span></p><p style="text-align:justify;line-height:150%;"><span style="font-family:Arial,Helvetica,sans-serif;font-size:small;"><span style="font-size:small;font-family:Arial,Helvetica,sans-serif;">The customary paradigm for the treatment of cancer is to induce ROS formation so that it causes irreversible oxidative damage. Our laboratory has shown that the most common molecular driver of AML, the FLT3 </span><span style="text-decoration:underline;">I</span><span style="font-size:small;font-family:Arial,Helvetica,sans-serif;">nternal </span><span style="text-decoration:underline;">T</span><span style="font-size:small;font-family:Arial,Helvetica,sans-serif;">andem </span><span style="text-decoration:underline;">D</span><span style="font-size:small;font-family:Arial,Helvetica,sans-serif;">uplication (ITD) mutation, leads to cells that produce very high levels of ROS that modify the bone marrow redox microenvironment. Further, using novel and sophisticated proteomics techniques we have shown that AML patients harbouring FLT-ITD+ mutations express proteins showing oxidation of catalytic cysteines in a number of key tumour suppressor proteins such as the protein tyrosine phosphatases (PTPs) which renders them inactive. These novel data may explain the hyperactive activated growth and survival signalling pathways that characterise this subtype of AML. </span></span></p><p style="text-align:justify;line-height:150%;"><span style="font-family:Arial,Helvetica,sans-serif;font-size:small;">The hypothesis for my project is that if we reduce intracellular ROS we may reduce the oxidation of a number of key tumour suppressor proteins we have shown to be inactivated in FLT3-ITD+ AMLs. Through the specific inhibition of ROS production using new FLT3 targeted therapies we will not only inhibit the oncogene responsible for transformation but we will reactivate the negative regulators of the FLT3 pathway, whilst reducing oxidative DNA damage that may help us to overcome issues with resistance and therefore reduce relapse rates and fatalities. </span></p><div><p style="text-align:justify;line-height:150%;"><span style="font-family:Arial,Helvetica,sans-serif;font-size:small;">In the studies described herein, we show that by inhibiting NOX2 we inhibit ROS production. When NOX2 inhibition is combined with FLT3 inhibition, we can induce synergistic cytotoxicity to FLT3-ITD+ AML cells when grown and treated in a model of the bone marrow characterised by low oxygen tension. We also present exciting preliminary mechanistic data showing that the FLT3 regulating PTPs, SHP-1 and DEP-1, are oxidised by ROS produced by NOX2, and that reduced oxidative stress through NOX2 inhibition increased PTP activity by reduced catalytic cysteine oxidation and increased PTP protein expression. </span></p><p style="text-align:justify;line-height:150%;"><span style="font-family:Arial,Helvetica,sans-serif;font-size:small;">The results of my project show NADPH oxidase as a potent source of reactive oxygen species that is responsible for high levels of oxidative stress seen in the most commonly occurring genetic mutation in AML – FLT3-ITD. ROS produced by the NADPH oxidase-2 complex causes increased DNA damage increasing the genomic instability of these cells and potentially influences the common resistance to therapies that we see. Additionally, my thesis sets the ground-work for a novel treatment paradigm that reduced oxidative stress, which leads to reactivation of intrinsic defence systems that work in cooperation with targeted therapies to synergistically kill AML cells. Finally, the impact of targeting the redox microenvironment in AML and the potential for modulating ROS production for the treatment of AML are discussed, along with the limitations and future directions of this study.</span></p></div><span style="font-size:12.0pt;line-height:150%;font-family:'Times New Roman',serif;"><br clear="all" style="page-break-before:always;" /></span> |
Medical Science, Faculty of Health and Medicine, University of Newcastle | Principal Supervisor |
| 2016 | Honours |
Prostate cancer: Quantification of proNGF by targeted proteomics Background. Prostate cancer (PCa) is one of the most prevalent cancers in Australia. The PCa serum biomarker prostate specific antigen (PSA) is used for screening, diagnosis and prognosis, but increasing evidence indicates that PSA testing has serious limitations. Not only does it lack specificity (with false positive and negative detections), but it also fails in identifying the most aggressive forms of PCa that require treatment as compared to the indolent forms of the disease. There is therefore an urgent need for new biomarkers to improve the diagnosis and prognosis of PCa. <br />Preliminary data from our lab have identified the precursor of nerve growth factor (proNGF) as a new potential serum biomarker for PCa. ProNGF expression is increased in cancer cells and positively correlates with PCa aggressiveness. In addition, PCa cells secrete proNGF, and by ELISA increased concentration of proNGF were found in the serum of PCa patients, in particular in case of aggressive tumours. These data suggest that proNGF could be a novel serum biomarker for diagnosis and prognosis of PCa. <br />Aims. This project is focused on developing a targeted proteomic assay (in parallel reaction monitoring - PRM) to reliably quantify proNGF in serum samples. The assay will subsequently be applied for the analysis of PCa serum samples. <br />Methods. After in silico trypsin digestion, two tryptic peptides unique to proNGF were selected with molecular mass and charge appropriate for detection in mass spectrometry. We have obtained the corresponding stable isotope labelled (SIL) peptides and we have established all experimental conditions for their detection in the serum and quantification in PRM mass spectrometry. In particular, we have analysed a series of PCa samples (n=40) of different grade and a series of benign prostatic hyperplasia (BPH, n=20). The results were compared with data obtained by ELISA for proNGF in the same samples. <br />Results. 1. An optimised PRM quantification approach was achieved by analysing twoproNGF SIL peptides LQHSLDTALR and SAPAAAIAAR (by direct injection and in complex serum background). 2. Standard curves for both proNGF SIL peptides in complex serum background were generated and performed to normalise inter-run variability of the mass spectrometer. 3. The average quantity of proNGF endogenous peptide LQHSLDTALR in serum samples was 0.021 ± 0.050 ng/mL, whereas proNGF endogenous peptide SAPAAAIAAR was undetectable. 4. No significant difference of proNGF peptide LQHSLDTALR quantity was found either between BPH and PCa cancer or across BPH group, low-risk PCa (Gleason 6) and high-risk PCa (Gleason 8 & 9). 5. The correlation factor (r) between PRM data for peptide LQHSLDTALR and the ELISA for proNGF was 0.21, indicating a limited correspondence between data obtained by the two methods. <br />Conclusion. We have developed a PRM based assay for quantifying proNGF in biological samples with a very good specificity/sensitivity (picogram/mL level). However, its use to quantify proNGF in the serum of PCa patients needs further improvements and in particular further optimisation in sample preparation, such as albumin depletion and deglycosylation. Such a PRM assay for proNGF could find biomedical applications beyond PCa, as proNGF has recently been reported to be increased in the cerebrospinal fluid of patients with neurodegenerative disease. <br />6 |
Medical Science, Faculty of Health and Medicine, University of Newcastle | Co-Supervisor |
| 2016 | Honours |
Development of a sensitive, robust and quantitative mass spectrometry assay for the measurement of activated signalling pathways in human breast cancers <p style="text-align:justify;text-justify:inter-ideograph;line-height:150%;"><span lang="EN-US" style="color:windowtext;">Breast cancer is a major national and global health concern. While the overall survival for breast cancer has significantly improved over the past decades, outcomes for patients with triple negative (TNBC) or Luminal B subtype tumuors remains poor. Recent studies have shown that some TNBCs display inactivation of key DNA damage repair (DDR) pathways which renders them hypersensitive to specific DNA damaging chemotherapy. Whether other breast cancer subtypes also have altered DDR pathways is not known, in part because an effective tool for quantifying the activity of DDR pathways is not available. Therefore, we aimed to develop a robust and sensitive assay to quantitate the activity of DDR pathways in breast cancer.</span><span style="color:windowtext;"> Recent studies indicate that </span><span style="color:windowtext;">a targeted proteomics approach, utilising </span><span style="color:windowtext;">quantitative mass spectrometry-based </span><span style="color:windowtext;">parallel reaction monitoring (PRM), may be a useful technique for identification and quantification of such pathways. PRM is a highly selective and specific method for targeted peptide quantification, including peptides with post-translational modifications such as phosphorylation. It is beginning to be implemented for the quantitative study of changes in protein components within cancer cells. Given that cancer associated signalling pathways are regulated by phosphorylation of key protein mediators, we hypothesized that</span><span style="color:windowtext;"> the development of</span><span style="color:windowtext;"> a multiplexed PRM assay for quantitation of key phosphoproteins involved in activated signalling pathways could be a powerful tool for the identification of activated, and potentially ‘druggable’ pathways in human breast tumours. </span></p><p style="text-align:justify;text-justify:inter-ideograph;line-height:150%;"><span>Sample preparation is the crucial starting point to obtain high-quality mass spectrometry data, therefore a major focus of this thesis was to determine the optimal sample preparation procedure for PRM-based quantitation of phospho-peptides. To do this,</span><span style="color:windowtext;"> the human breast cancer cell line, </span><span lang="EN-US" style="color:windowtext;">BT474, was </span><span style="color:windowtext;">treated +/- bleomycin to induce double stranded DNA breaks (DSBs) and activate DSB repair pathways. Proteins were extracted in either a sodium carbonate buffer or via the commercially available </span><span lang="EN-US">ProteaseMax<sup>TM</sup></span><span style="color:windowtext;"> kit, and enzymatically digested into peptides using trypsin and lysozyme C, and identified by liquid chromatography tandem mass spectrometry (LC-MS/MS). Analysis of the identified peptides demonstrated </span><span lang="EN-US">enhanced tryptic/Lys C digestion efficiency with the sodium carbonate method, and as such this was used for all subsequent experiments. However, phosphorylated peptides only constituted ~1% of the total peptides identified, and the peak intensity for most phospho-peptides was low, making accurate quantitation difficult. Recent studies have shown <a name="OLE_LINK41">enrichment of phosphopeptides</a> using titanium dioxide chromatography (TiO2) significantly increases the number of phosphoproteins identified in discovery MS projects. Whether phospho-peptide enrichment is required for targeted PRM MS approaches however has not been determined. Therefore, a direct comparison of non-enriched versus phospho-peptide enrichment using a range of TiO2 procedures were performed in the BT474 cells. This resulted in high abundance of non-modified peptides in the non-enriched and flow through lysates, while the majority of phosphopeptides were found in TiO2 eluates. The enrichment enabled improved quantitation of specific phosphopeptides by PRM, and thus confirmed that enrichment with at least one TiO2 step was required for quantitative detection of phosphopeptides. </span></p><p style="text-align:justify;text-justify:inter-ideograph;line-height:150%;"><span lang="EN-US">This optimized procedure was then used to quantitate key phosphoproteins involved in growth, survival and DNA damage repair pathways in breast cancer. </span><span style="color:windowtext;">As a positive control we analysed H2AX, a key marker of DSBs, and as expected found increased phosphorylation of pSer139-H2AX in response to bleomycin. We further found increased phosphorylation of </span><span lang="EN-US" style="color:windowtext;">Protein Kinase B (AKT; Thr307 and Ser473), </span><span style="color:windowtext;">checkpoint kinase 2 (CHK2) at Thr386, and the ataxia-telangiectasia mutated (ATM) protein (double phosphorylation at Y175 and S179). In contrast H2AX doubly phosphorylated at S139/Y140, pThr68-CHK2 and pSer367-ATM was reduced. This provides novel insight into the cellular response to DSBs. Importantly, t</span><span lang="EN-US" style="color:windowtext;">his analysis was achieved from average 2 µg of peptides, therefore we believe this assay will be useful for primary patient tissues and hence may be a powerful approach for identifying activated and druggable pathways in human breast tumuors.</span></p> |
Medical Science, Faculty of Health and Medicine, University of Newcastle | Co-Supervisor |
| 2015 | Honours |
The Role of Reduced Protein Phosphatase 2A Subunit, PPP2R2A (B55α), Expression in Breast Cancer. <p style="margin-bottom:0cm;margin-bottom:.0001pt;line-height:150%;"><span style="font-size:12.0pt;line-height:150%;font-family:'Times New Roman',serif;">Breast Cancer is a disease of global concern, with current treatments lacking selectivity and not showing a response in many patients. Patients with luminal-B like breast cancers in particular have shown reduced sensitivity to endocrine therapies, and have a more aggressive phenotype similar to Triple Negative breast tumours and worse prognosis than luminal-A like breast tumours. A marker and potential therapeutic target was thus need to distinguish these particular patients, as current methods of classification have been shown to be ineffective, especially in distinguishing between luminal-A and B tumours. Recent studies have shown that reduced expression of the <em>PPP2R2A</em> gene, encoding the B55a subunit of Protein Phosphatase 2a (PP2A), may be such a marker, with highly reduced expression in a majority of luminal-B tumours studied. Reduced expression of this subunit has been found to impair the DNA damage Homologous Recombination (HR) repair pathway in HeLa cervical cancer cells, and furthermore, reduced B55a expression and impaired DNA damage repair in lung carcinomas was found to sensitise these cells to the PARP inhibitor (PARPi) ABT888, through synthetic lethality. </span></p><p style="margin-bottom:0cm;margin-bottom:.0001pt;line-height:150%;"><span style="font-size:12.0pt;line-height:150%;font-family:'Times New Roman',serif;"></span></p><p style="margin-bottom:0cm;margin-bottom:.0001pt;line-height:150%;"><span style="font-size:12.0pt;line-height:150%;font-family:'Times New Roman',serif;">This study therefore investigated the role reduced B55a expression may play in breast cancer cells, to determine whether this is a potential treatment target for low-B55a expressing luminal-B tumours. It was first hypothesised, based on past studies, that reduced B55a expression in breast cancer cells would impair their DNA damage repair pathways. To investigate this, two B55a knockdown models were created in both<span>&nbsp; </span>MCF7 and ZR751breast cancer cell lines, as well as a control cell line to compare back to. DNA repair efficiency was then examined by inducing DNA damage via Bleomycin and analysing expression of important HR repair proteins. Changes in expression and phosphorylation mediated activation of many of these proteins, as well as prolonged increase of ɣH2AX (marker for DSBs) in B55a-low cells compared to control cells supported the hypothesis that DN repair was impaired in these B55a-low cells. </span></p><p style="margin-bottom:0cm;margin-bottom:.0001pt;line-height:150%;"><span style="font-size:12.0pt;line-height:150%;font-family:'Times New Roman',serif;"></span></p><p style="margin-bottom:0cm;margin-bottom:.0001pt;line-height:150%;"><span style="font-size:12.0pt;line-height:150%;font-family:'Times New Roman',serif;">It was then hypothesised that impaired DNA repair in B55a-low breast cancer cells would further sensitise these cells to DNA damaging agents. To examine this, B55a knockdown and control cells were treated with DNA damaging agents (ɣ-irradiation, Bleomycin, Cisplatin, ABT888), at various doses. However the hypothesis was not supported, as there was no significantly increased sensitivity to these agents in B55a-low cells compared to the control cells.</span></p><p style="margin-bottom:0cm;margin-bottom:.0001pt;line-height:150%;"><span style="font-size:12.0pt;line-height:150%;font-family:'Times New Roman',serif;"></span></p><span style="font-size:12.0pt;line-height:107%;font-family:'Times New Roman',serif;">Therefore while this study suggests that DNA repair may be altered and impaired in B55a-low breast cancer cells (and hence luminal-B tumours), this finding is currently not translatable to use as a potential treatment target for patient with luminal-B tumours. However future studies using altered dosage and time course of DNA damaging agents may find a heightened sensitivity of B55a-low breast cancer cell lines, allowing for specific personalised treatment of patients with aggressive luminal-B tumours, using therapeutics that already currently available. </span> |
Medical Science, Faculty of Health and Medicine, University of Newcastle | Co-Supervisor |
| 2014 | Honours |
Investigation of the function of tetraspanins CD9 and CD151 on prostate exosomes Exosomes are nanosized, membranous vesicles secreted by most cells that have been shown to be important for cellular communication. Whilst most research into exosomes has focussed on their function in a diseased state, such as cancer, their role in normal cellular functioning has not been thoroughly investigated. Tetraspanins are a family of transmembrane proteins, including CD9 and CD151, that are abundantly expressed on exosomes, however, their function within exosomes is yet to be fully elucidated. CD9 and CD151 have opposing roles in many solid tumours, including prostate, where CD9 is typically downregulated and CD151 is typically upregulated and both being indicative of poor prognosis. Therefore, this study was designed to investigate the effects that altered CD9 and CD151 in exosomes have on a normal cellular population, and whether alteration of these tetraspanins can make a normal cell more metastatic.<br />CD9 and CD151 expression was altered in a normal, non-tumourigenic cell line (RWPE1) to express either low CD9 (CD9 low) or high CD151 (CD151 high). Western blot and flow cytometry were used to confirm that these cell lines displayed altered expression of CD9 and CD151 and that their control lines (Scram and Zeo respectively) displayed similar expression to RWPE1 cells. An invasive and metastatic derivative cell line of RWPE1 cells, WPE1-NB26, was used as a tumourigenic control. Exosomes from these cells were then assessed for their ability to attach to and internalise into RWPE1 cells before being used as treatments for RWPE1 cells in a range of functional assays, including adhesion, proliferation, migration and invasion, to assess whether the altered exosomal expression of CD9 and CD151 can influence these cellular behaviours. Exosomes from RWPE1, CD9 low, CD151 high and WPE1-NB26 cell lines were also analysed using a quantitative mass spectroscopy approach to determine changes in their proteome.<br />There were no differences observed between exosome populations in regards to attachment and internalisation into RWPE1 cells. Treating RWPE1 cells with exosomes also showed no difference in the proliferation of RWPE1 cells over a 72<br />6<br />hour period. RWPE1 cell adhesion to Collagen-IV, Fibronectin and Laminin 1 was assessed and revealed no difference for CD9 low exosome treated cells, however, an increase in adhesion was seen with the CD151 high exosome treated cells at 1 (p = 0.0023) and 4 hours (p = 0.0279). Only CD151 high exosome treated RWPE1 cells showed an increase in migration at 20 (p = 0.0186), 24 (p = 0.0068) and 30 hours (p = 0.0038), however no differences were observed in an invasion assay. Proteomic analysis of exosomes revealed that there were 101 proteins with a greater than 2 fold increase and 18 proteins with a greater than 2 fold decrease in abundance compared to RWPE1 exosomes.<br />The results of this study suggest that alterations of CD9 and CD151 in exosomes have only limited functional effects on a normal cellular population. This study also identified that altering the expression of CD9 and CD151 in exosomes affects their entire proteome. Further characterisation of the effects of CD9 and CD151 expression in exosomes needs to be performed, utilising a panel of tumourigenic cell lines. Therefore due to the alterations on cellular function upon the addition of CD151 high exosomes, it suggests that modified exosomes could represent a therapeutic strategy for prostate cancer. |
Medical Science, Faculty of Health and Medicine, University of Newcastle | Co-Supervisor |
Research Collaborations
The map is a representation of a researchers co-authorship with collaborators across the globe. The map displays the number of publications against a country, where there is at least one co-author based in that country. Data is sourced from the University of Newcastle research publication management system (NURO) and may not fully represent the authors complete body of work.
| Country | Count of Publications | |
|---|---|---|
| Australia | 168 | |
| United States | 51 | |
| Netherlands | 25 | |
| Switzerland | 19 | |
| Denmark | 19 | |
| More... | ||
News
News • 19 Apr 2024
Newcastle team on mission to improve childhood cancer outcomes
A University of Newcastle and Hunter Medical Research Institute team will endeavour to improve the outcomes of children diagnosed with high-risk cancers with support of a $591,890 Cancer Australia grant.
News • 9 Jun 2023
Symposium spurs race to beat brain cancer
Experts from across the nation are tackling brain cancer head on with collaboration front and centre following the inaugural symposium of the University of Newcastle’s Mark Hughes Foundation Centre for Brain Cancer Research.
News • 30 May 2023
Drug combination discovery extending life expectancy of children with brain cancer
Newcastle and Hunter-based researchers have discovered a drug combination that could dramatically improve the life expectancy of children living with brain cancer.
News • 29 Mar 2023
Discovery key to reducing leukaemia treatment resistance
In a world first, Newcastle researchers have discovered the mechanisms acute myeloid leukaemia (AML) cells use to produce ‘free radicals’ – the byproduct of a cell process that aggressively fuels the growth of cancer cells and limits the effectiveness of current treatments.
News • 12 Mar 2023
Associate Professor Matt Dun named Lake Mac Ambassador for 2023
University of Newcastle cancer researcher, Associate Professor Matt Dun, has been selected as Lake Macquarie’s Ambassador of the Year for 2023.
News • 2 Nov 2020
New treatment idea discovered for leukaemia
Newcastle researchers have discovered a new way to kill leukaemia cells.
News • 12 Aug 2020
Newcastle cancer researcher recognised as Young Tall Poppy
Esteemed cancer researcher and charity advocate, Dr Matthew Dun, has been acknowledged for his dedication to improving cancer survival outcomes, with a 2020 ‘Young Tall Poppy’ science award.
News • 20 Jul 2020
Tests show potential for medicinal cannabis to kill cancer cells
Laboratory tests conducted at the University of Newcastle and Hunter Medical Research Institute have shown that a modified form of medicinal cannabis can kill or inhibit cancer cells without impacting normal cells, revealing its potential as a treatment rather than simply a relief medication.
News • 6 Nov 2019
Biomedical scientist’s mission to conquer cancer recognised at NSW Premier's Awards
Dr Matt Dun was named the Outstanding Cancer Research Fellow at this year's NSW's Premier Awards for Outstanding Cancer Research.
News • 30 Aug 2019
$10 million funding boost for local health researchers
More than $10 million has been awarded to further world-class, solution-based research at the University of Newcastle, Hunter Medical Research Institute (HMRI), and Hunter New England Health in the latest round of National Health and Medical Research Council (NHMRC) funding.
News • 25 Sep 2017
Strengthening the defence against Leukaemia
Research from the University of Newcastle (UON) has revealed a more targeted way of treating T-cell acute lymphoblastic leukaemia (T-ALL), an aggressive type of cancer typically prevalent in children and adolescents.
News • 21 Feb 2014
$1.5m to Newcastle researchers to unlock cancer secrets
Three University of Newcastle cancer researchers have been recognised with Cancer Institute NSW Early Career Fellowships, totalling more than $1.5 million.
Professor Matt Dun
Position
Professor
Molecular Oncology
School of Biomedical Sciences and Pharmacy
College of Health, Medicine and Wellbeing
Contact Details
| matt.dun@newcastle.edu.au | |
| Phone | (02) 4921 5693 |
| Fax | (02) 4921 6903 |
| Link |
Office
| Room | LS3.33. |
|---|---|
| Building | Life Sciences Building |
| Location | Callaghan University Drive Callaghan, NSW 2308 Australia |